CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.788 | 59th percentile | |
Fraction of mapped reads for the background samples: |
0.761 | 57th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.245 | 70th percentile | |
Error in fit of enrichment distribution: |
0.406 | 21st percentile | |
Peak statistics |
|||
Number of peaks: |
19318 | 61st percentile | |
Fraction of reads mapped to peaks: |
0.026 | 55th percentile | |
Motif enrichment statistics |
|||
Enrichment of top motif: |
1.76 | 15th percentile | |
Enrichment of complimentary motif set: |
3.21 | 10th percentile | |
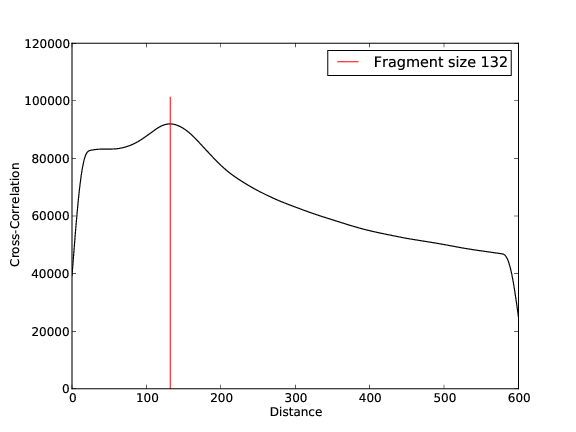
Quality Control, Mapping, Fragment Size Estimation:
BCLAF Replicate 1
- 31,159,630 input reads (in FASTQ format).
- 25,734,688 reads after removal of low quality reads.
- 25,596,580 reads after removal of adapters and low complexity reads.
- 24,269,988 reads after mapping.
- Report PDF
BCLAF Replicate 2
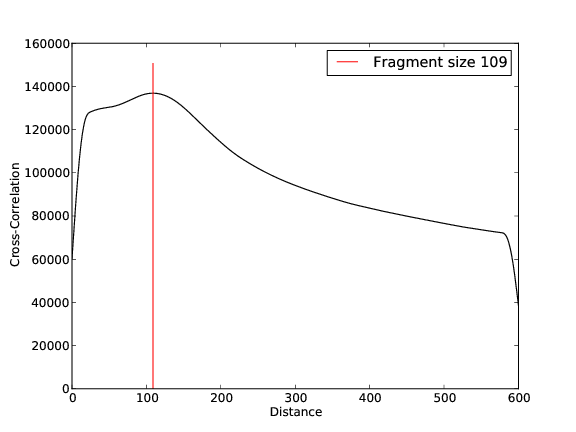
- 37,615,056 input reads (in FASTQ format).
- 32,932,491 reads after removal of low quality reads.
- 32,656,957 reads after removal of adapters and low complexity reads.
- 29,914,587 reads after mapping.
- Report PDF
BG Replicate 1
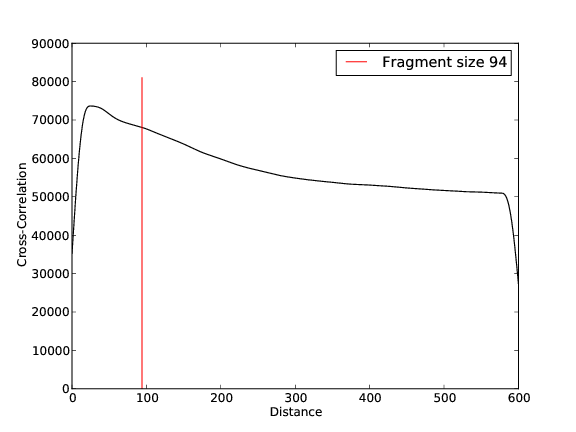
- 52,452,124 input reads (in FASTQ format).
- 48,183,574 reads after removal of low quality reads.
- 47,412,139 reads after removal of adapters and low complexity reads.
- 36,430,732 reads after mapping.
- Report PDF
BG Replicate 2
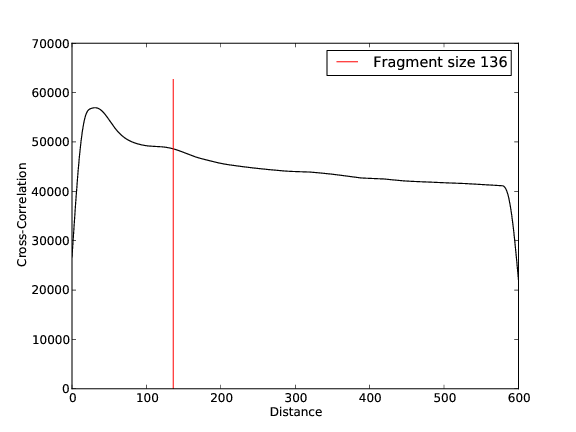
- 62,063,640 input reads (in FASTQ format).
- 54,936,648 reads after removal of low quality reads.
- 54,705,378 reads after removal of adapters and low complexity reads.
- 50,697,602 reads after mapping.
- Report PDF
Peak Calling:
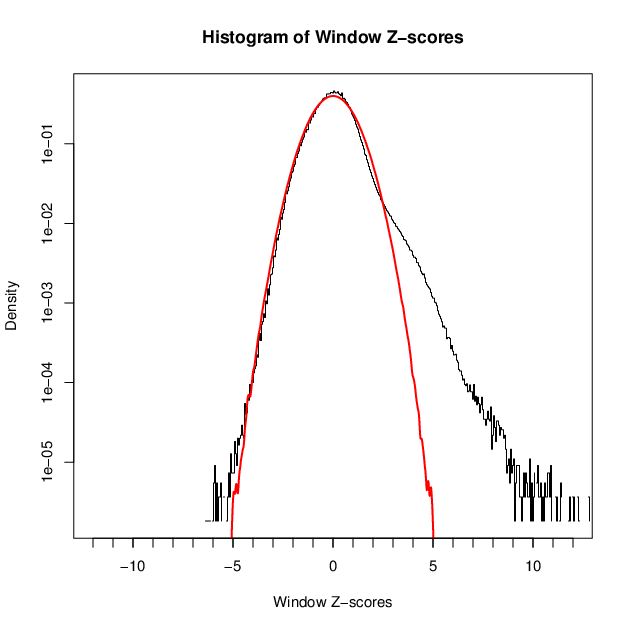
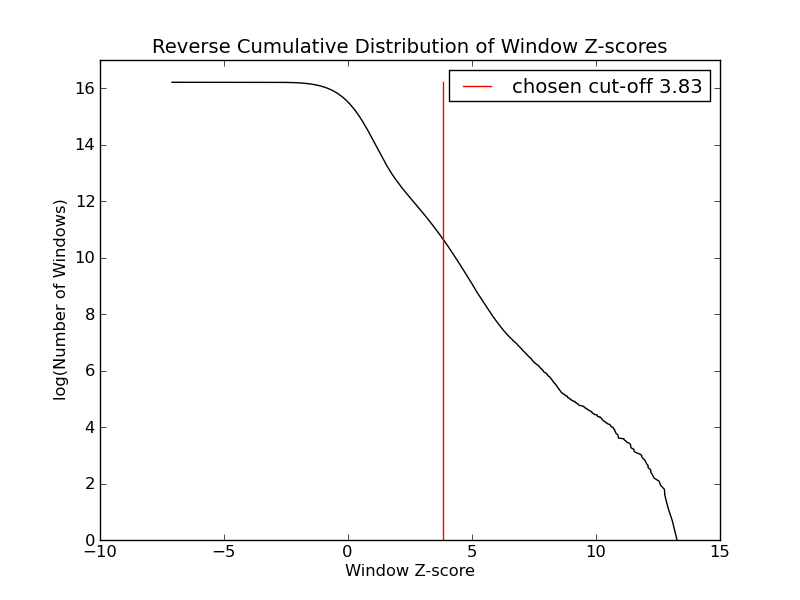
- 42526 windows are significantly enriched at FDR=10% (z-value cut-off: 3.83)
- 42526 windows merged to 21819 regions.
- 19318 significant peaks fitted within 21819 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | HCMC.ELF1_f1.wm | HCMC.IRF8_si.wm | ZNF143.p2 | denovo_WM_17 | HCMC.CTCF_f2.wm | denovo_WM_21 | HCMC.SPI1_si.wm | JASPAR.Myf.wm | HCMC.FOXM1_f1.wm |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| chr10:103124593..103124696 | 12.491 | BTRC|beta-transducin repeat containing | -10772 (+) | POLL|polymerase (DNA directed), lambda | 223285 (-) | |||||||||
| chr14:35025372..35025563 | 12.490 | EAPP|E2F-associated phosphoprotein | -16582 (-) | SNX6|sorting nexin 6 | 73888 (-) | |||||||||
| chr14:35015953..35016144 | 12.461 | EAPP|E2F-associated phosphoprotein | -7163 (-) | SNX6|sorting nexin 6 | 83307 (-) | |||||||||
| chr17:19091361..19091525 | 12.361 | GRAPL|GRB2-related adaptor protein-like | -60662 (+) | EPN2|epsin 2 | 49265 (+) | -30:0.293 |
29:0.283 |
|||||||
| chr17:18967212..18967403 | 12.164 | GRAP|GRB2-related adaptor protein | -17021 (-) | GRAPL|GRB2-related adaptor protein-like | 63474 (+) | 59:0.293 |
0:0.283 |
|||||||
| chr17:8076704..8076895 | 11.858 | VAMP2|vesicle-associated membrane protein 2 (synaptobrevin 2) | -10507 (-) | TMEM107|transmembrane protein 107 | 2861 (-) | |||||||||
| chr1:45187349..45187540 | 11.566 | C1orf228|chromosome 1 open reading frame 228 | -47051 (+) | KIF2C|kinesin family member 2C | 18054 (+) | |||||||||
| chr1:147511007..147511160 | 11.539 | GPR89C|G protein-coupled receptor 89C | -85517 (+) | NBPF11|neuroblastoma breakpoint family, member 11 NBPF24|neuroblastoma breakpoint family, member 24 | 98997 (-) | |||||||||
| chr15:65588392..65588576 | 11.523 | PARP16|poly (ADP-ribose) polymerase family, member 16 | -9324 (-) | IGDCC3|immunoglobulin superfamily, DCC subclass, member 3 | 81867 (-) | 58:0.256 |
||||||||
| chr1:145969276..145969426 | 11.495 | GPR89C|G protein-coupled receptor 89C | -85484 (+) | NBPF11|neuroblastoma breakpoint family, member 11 NBPF24|neuroblastoma breakpoint family, member 24 | 98893 (-) | |||||||||
| chr17:19093312..19093495 | 11.402 | GRAPL|GRB2-related adaptor protein-like | -62622 (+) | EPN2|epsin 2 | 47305 (+) | 72:0.293 |
12:0.283 |
|||||||
| chr17:18965367..18965537 | 11.332 | GRAP|GRB2-related adaptor protein | -15166 (-) | GRAPL|GRB2-related adaptor protein-like | 65329 (+) | 45:0.509 |
||||||||
| chr22:50338926..50339103 | 11.289 | CRELD2|cysteine-rich with EGF-like domains 2 | -26639 (+) | PIM3|pim-3 oncogene | 15135 (+) | 1:0.315, -42:0.315, 46:0.315, -86:0.315, 91:0.315 |
||||||||
| chr12:120729515..120729706 | 11.281 | PXN|paxillin | -26020 (-) | SIRT4|sirtuin 4 | 10532 (+) | |||||||||
| chr1:11968216..11968407 | 11.273 | NPPB|natriuretic peptide B | -49371 (-) | KIAA2013|KIAA2013 | 18162 (-) | |||||||||
| chr17:19015676..19015867 | 11.266 | GRAP|GRB2-related adaptor protein | -65485 (-) | GRAPL|GRB2-related adaptor protein-like | 15010 (+) | -93:0.509, 95:0.293 |
35:0.283 |
|||||||
| chr17:41438490..41438681 | 11.125 | TMEM106A|transmembrane protein 106A | -74706 (+) | LOC100130581|uncharacterized LOC100130581 | 27105 (-) | |||||||||
| chr1:147994170..147994360 | 11.052 | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | -38868 (-) | NBPF14|neuroblastoma breakpoint family, member 14 NBPF1|neuroblastoma breakpoint family, member 1 | 31589 (-) | |||||||||
| chr1:148241480..148241613 | 10.828 | PPIAL4D|peptidylprolyl isomerase A (cyclophilin A)-like 4D PPIAL4F|peptidylprolyl isomerase A (cyclophilin A)-like 4F | -39011 (-) | NBPF15|neuroblastoma breakpoint family, member 15 | 316681 (+) | |||||||||
| chr1:28975108..28975276 | 10.745 | TAF12|TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa | -5612 (-) | GMEB1|glucocorticoid modulatory element binding protein 1 | 20037 (+) | 26:0.691 |
||||||||
| chr13:114103532..114103717 | 10.717 | ADPRHL1|ADP-ribosylhydrolase like 1 | -170 (-) | ADPRHL1|ADP-ribosylhydrolase like 1 | 4214 (-) | -9:0.293, 24:0.292, 58:0.293, -76:0.293, 91:0.293, -110:0.293 |
-74:0.261, 93:0.261, -108:0.261 |
|||||||
| chr2:3622714..3622828 | 10.680 | RNASEH1|ribonuclease H1 | -16829 (-) | 8 (-) | 21:0.754 |
|||||||||
| chr1:149514074..149514238 | 10.639 | HIST2H2BF|histone cluster 2, H2bf | -114960 (-) | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | 38867 (+) | |||||||||
| chr8:56987084..56987232 | 10.381 | RPS20|ribosomal protein S20 | -72 (-) | MOS|v-mos Moloney murine sarcoma viral oncogene homolog | 39382 (-) | -23:0.517 |
||||||||
| chr17:56709001..56709186 | 10.355 | C17orf47|chromosome 17 open reading frame 47 | -87386 (-) | TEX14|testis expressed 14 | 60302 (-) | |||||||||
| chr17:56736476..56736667 | 10.334 | C17orf47|chromosome 17 open reading frame 47 | -114864 (-) | TEX14|testis expressed 14 | 32824 (-) | |||||||||
| chr12:38710443..38710582 | 10.317 | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | -4535039 (+) | ALG10B|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast) | 113 (+) | 3:0.299, 56:0.328, -76:0.620 |
43:0.923 |
|||||||
| chr1:148604852..148605039 | 10.273 | NBPF15|neuroblastoma breakpoint family, member 15 NBPF16|neuroblastoma breakpoint family, member 16 | -27529 (+) | PPIAL4E|peptidylprolyl isomerase A (cyclophilin A)-like 4E | 39065 (+) | |||||||||
| chr17:56756866..56757049 | 10.263 | C17orf47|chromosome 17 open reading frame 47 | -135250 (-) | TEX14|testis expressed 14 | 12438 (-) | |||||||||
| chr18:9841..10032 | 10.251 | None (None) | USP14|ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | 148594 (+) | ||||||||||
| chr16:89307471..89307659 | 10.249 | ZNF778|zinc finger protein 778 | -23438 (+) | ANKRD11|ankyrin repeat domain 11 | 42509 (-) | -3:0.744, 26:0.744, -33:0.744, 56:0.744, -63:0.744, 86:0.744, -93:0.744, 116:0.744 |
||||||||
| chr1:11969787..11969943 | 10.248 | NPPB|natriuretic peptide B | -50925 (-) | KIAA2013|KIAA2013 | 16608 (-) | |||||||||
| chr1:45196644..45196834 | 10.216 | C1orf228|chromosome 1 open reading frame 228 | -56346 (+) | KIF2C|kinesin family member 2C | 8759 (+) | |||||||||
| chr17:18965281..18965359 | 10.159 | GRAP|GRB2-related adaptor protein | -15034 (-) | GRAPL|GRB2-related adaptor protein-like | 65461 (+) | -11:0.293 |
48:0.283 |
|||||||
| chr8:143751378..143751483 | 10.152 | JRK|jerky homolog (mouse) | -29 (-) | PSCA|prostate stem cell antigen | 10476 (+) | 10:0.774, -48:0.896 |
||||||||
| chr1:149605958..149606137 | 10.105 | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | -53024 (+) | FCGR1A|Fc fragment of IgG, high affinity Ia, receptor (CD64) | 148225 (+) | |||||||||
| chr15:65597073..65597244 | 10.098 | PARP16|poly (ADP-ribose) polymerase family, member 16 | -17998 (-) | IGDCC3|immunoglobulin superfamily, DCC subclass, member 3 | 73193 (-) | |||||||||
| chr1:146556130..146556321 | 10.049 | NBPF11|neuroblastoma breakpoint family, member 11 NBPF24|neuroblastoma breakpoint family, member 24 | -473527 (-) | PRKAB2|protein kinase, AMP-activated, beta 2 non-catalytic subunit | 87540 (-) | 92:0.535 |
||||||||
| chr3:73160038..73160170 | 9.990 | EBLN2|endogenous Bornavirus-like nucleoprotein 2 | -49295 (+) | PDZRN3|PDZ domain containing ring finger 3 | 513934 (-) | |||||||||
| chr1:148241615..148241804 | 9.986 | PPIAL4D|peptidylprolyl isomerase A (cyclophilin A)-like 4D PPIAL4F|peptidylprolyl isomerase A (cyclophilin A)-like 4F | -39174 (-) | NBPF15|neuroblastoma breakpoint family, member 15 | 316518 (+) | |||||||||
| chr1:9991..10182 | 9.961 | None (None) | WASH2P|WAS protein family homolog 2 pseudogene WASH3P|WAS protein family homolog 3 pseudogene WASH7P|WAS protein family homolog 7 pseudogene | 19319 (-) | ||||||||||
| chr1:38455703..38455849 | 9.950 | SF3A3|splicing factor 3a, subunit 3, 60kDa | -78 (-) | FHL3|four and a half LIM domains 3 | 15457 (-) | -15:0.896, -91:0.643 |
61:0.994, 114:0.643 |
64:0.679, -85:0.338 |
||||||
| chr1:1310586..1310706 | 9.895 | AURKAIP1|aurora kinase A interacting protein 1 | -73 (-) | AURKAIP1|aurora kinase A interacting protein 1 | 217 (-) | -64:0.912, 68:0.358 |
||||||||
| chr2:224822085..224822198 | 9.860 | WDFY1|WD repeat and FYVE domain containing 1 | -12076 (-) | MRPL44|mitochondrial ribosomal protein L44 | 12 (+) | |||||||||
| chr7:141437993..141438136 | 9.859 | WEE2|WEE1 homolog 2 (S. pombe) | -29912 (+) | SSBP1|single-stranded DNA binding protein 1 | 200 (+) | 102:0.896 |
57:0.276 |
|||||||
| chr4:106629830..106629947 | 9.712 | INTS12|integrator complex subunit 12 | -47 (-) | GSTCD|glutathione S-transferase, C-terminal domain containing | 85 (+) | 45:0.261, 67:0.233 |
||||||||
| chr1:143647004..143647126 | 9.704 | -179440 (-) | FLJ39739|uncharacterized FLJ39739 LOC100286793|uncharacterized LOC100286793 | 97340 (-) | ||||||||||
| chr21:26950692..26950883 | 9.676 | MIR155HG|MIR155 host gene (non-protein coding) | -16315 (+) | MRPL39|mitochondrial ribosomal protein L39 | 29028 (-) | -112:0.911 |
||||||||
| chr1:147834492..147834613 | 9.640 | NBPF24|neuroblastoma breakpoint family, member 24 | -209952 (-) | FLJ39739|uncharacterized FLJ39739 | 97376 (-) | |||||||||
| chr14:35016190..35016381 | 9.516 | EAPP|E2F-associated phosphoprotein | -7400 (-) | SNX6|sorting nexin 6 | 83070 (-) | 99:0.379, -106:0.336 |
||||||||
| chr14:35025132..35025323 | 9.497 | EAPP|E2F-associated phosphoprotein | -16342 (-) | SNX6|sorting nexin 6 | 74128 (-) | -95:0.379, 110:0.336 |
||||||||
| chr1:148766903..148767094 | 9.493 | NBPF15|neuroblastoma breakpoint family, member 15 NBPF16|neuroblastoma breakpoint family, member 16 NBPF8|neuroblastoma breakpoint family, member 8 | -27497 (+) | PPIAL4D|peptidylprolyl isomerase A (cyclophilin A)-like 4D PPIAL4F|peptidylprolyl isomerase A (cyclophilin A)-like 4F | 39016 (+) | |||||||||
| chr2:122288448..122288638 | 9.447 | CLASP1|cytoplasmic linker associated protein 1 | -40624 (-) | CLASP1|cytoplasmic linker associated protein 1 | 118479 (-) | -117:0.779 |
42:0.257 |
|||||||
| chr1:45285491..45285681 | 9.406 | BTBD19|BTB (POZ) domain containing 19 | -11376 (+) | PTCH2|patched 2 | 23051 (-) | -22:0.318, 94:0.541 |
||||||||
| chr1:9798..9919 | 9.406 | None (None) | WASH2P|WAS protein family homolog 2 pseudogene WASH3P|WAS protein family homolog 3 pseudogene WASH7P|WAS protein family homolog 7 pseudogene | 19547 (-) | ||||||||||
| chr1:147806722..147806888 | 9.396 | NBPF24|neuroblastoma breakpoint family, member 24 | -182205 (-) | FLJ39739|uncharacterized FLJ39739 | 125123 (-) | |||||||||
| chr1:145382834..145382997 | 9.385 | NBPF10|neuroblastoma breakpoint family, member 10 NBPF8|neuroblastoma breakpoint family, member 8 | -89614 (+) | HFE2|hemochromatosis type 2 (juvenile) | 30348 (+) | |||||||||
| chr17:41393253..41393444 | 9.360 | TMEM106A|transmembrane protein 106A | -29469 (+) | LOC100130581|uncharacterized LOC100130581 | 72342 (-) | |||||||||
| chr18:10082..10273 | 9.360 | None (None) | USP14|ubiquitin specific peptidase 14 (tRNA-guanine transglycosylase) | 148353 (+) | ||||||||||
| chr1_gl000192_random:544946..545096 | 9.352 | None | None (None) | None | None (None) | |||||||||
| chr2:242041713..242041847 | 9.338 | MTERFD2|MTERF domain containing 2 | -54 (-) | PASK|PAS domain containing serine/threonine kinase | 47091 (-) | -27:0.896, -56:0.644, 109:0.289 |
||||||||
| chr1:144534125..144534305 | 9.335 | FAM91A2|family with sequence similarity 91, member A2 | -12986 (-) | NBPF10|neuroblastoma breakpoint family, member 10 | 59483 (+) | |||||||||
| chr17:30771379..30771510 | 9.312 | ZNF207|zinc finger protein 207 | -94199 (+) | PSMD11|proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | 45 (+) | 53:0.354 |
-18:0.954 |
-109:0.886 |
||||||
| chr6:89773263..89773342 | 9.301 | RNGTT|RNA guanylyltransferase and 5'-phosphatase | -99955 (-) | PNRC1|proline-rich nuclear receptor coactivator 1 | 17135 (+) | |||||||||
| chr15:49913173..49913297 | 9.243 | C15orf33|chromosome 15 open reading frame 33 | -152 (-) | DTWD1|DTW domain containing 1 | 14 (+) | -32:0.390 |
||||||||
| chr15:64679902..64680055 | 9.242 | KIAA0101|KIAA0101 | -6343 (-) | TRIP4|thyroid hormone receptor interactor 4 | 58 (+) | -83:0.429, -102:0.264 |
||||||||
| chr1:205091360..205091478 | 9.235 | RBBP5|retinoblastoma binding protein 5 | -272 (-) | DSTYK|dual serine/threonine and tyrosine protein kinase | 89254 (-) | 15:0.203 |
||||||||
| chr1:40723777..40723914 | 9.181 | ZMPSTE24|zinc metallopeptidase (STE24 homolog, S. cerevisiae) | -69 (+) | ZMPSTE24|zinc metallopeptidase (STE24 homolog, S. cerevisiae) | 1 (+) | |||||||||
| chr2:74682029..74682197 | 9.176 | RTKN|rhotekin | -13048 (-) | INO80B|INO80 complex subunit B | 91 (+) | 28:0.825 |
||||||||
| chr17:19091579..19091770 | 9.151 | GRAPL|GRB2-related adaptor protein-like | -60893 (+) | EPN2|epsin 2 | 49034 (+) | 12:0.240 |
||||||||
| chr17:41445248..41445439 | 9.141 | TMEM106A|transmembrane protein 106A | -81464 (+) | LOC100130581|uncharacterized LOC100130581 | 20347 (-) | |||||||||
| chr1:144311193..144311351 | 9.136 | FAM72D|family with sequence similarity 72, member D | -398142 (-) | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | 52952 (-) | |||||||||
| chr12:120729320..120729510 | 9.129 | PXN|paxillin | -25825 (-) | SIRT4|sirtuin 4 | 10727 (+) | |||||||||
| chr12:120730885..120731066 | 9.116 | PXN|paxillin | -27385 (-) | SIRT4|sirtuin 4 | 9167 (+) | |||||||||
| chr1:144491423..144491580 | 9.107 | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | -127277 (-) | LOC728855|uncharacterized LOC728855 | 29430 (-) | |||||||||
| chr12:34175297..34175429 | 9.103 | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | -148 (+) | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | 23 (+) | 45:0.328, -88:0.560 |
32:0.923 |
|||||||
| chr1:205180810..205180927 | 9.054 | DSTYK|dual serine/threonine and tyrosine protein kinase | -149 (-) | TMCC2|transmembrane and coiled-coil domain family 2 | 16215 (+) | 49:0.990 |
||||||||
| chr1:999654..999845 | 9.044 | LOC100288175|uncharacterized LOC100288175 | -4637 (+) | RNF223|ring finger protein 223 | 9937 (-) | -91:0.602 |
-120:0.240 |
|||||||
| chr19:59031191..59031381 | 9.012 | ZBTB45|zinc finger and BTB domain containing 45 | -98 (-) | TRIM28|tripartite motif containing 28 | 24546 (+) | -31:0.588, 90:0.647, -106:0.882 |
||||||||
| chr21:26950267..26950458 | 9.011 | MIR155HG|MIR155 host gene (non-protein coding) | -15890 (+) | MRPL39|mitochondrial ribosomal protein L39 | 29453 (-) | -40:0.310 |
||||||||
| chr11:64863542..64863672 | 9.001 | LOC100130348|uncharacterized LOC100130348 LOC399904|uncharacterized LOC399904 | -7048 (-) | C11orf2|chromosome 11 open reading frame 2 | 38 (+) | 55:0.880, -92:0.313 |
||||||||
| chrX:73164042..73164173 | 8.972 | CHIC1|cysteine-rich hydrophobic domain 1 | -380881 (+) | JPX|JPX transcript, XIST activator (non-protein coding) | 62 (+) | -46:0.389, -63:0.436, -80:0.434, -97:0.434 |
||||||||
| chr21:26951019..26951210 | 8.958 | MIR155HG|MIR155 host gene (non-protein coding) | -16642 (+) | MRPL39|mitochondrial ribosomal protein L39 | 28701 (-) | |||||||||
| chr22:23101293..23101484 | 8.953 | IGLJ3|immunoglobulin lambda joining 3 | -38257 (+) | 33718 (+) | ||||||||||
| chr14:61201380..61201514 | 8.874 | SIX4|SIX homeobox 4 | -10390 (-) | MNAT1|menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) | 68 (+) | |||||||||
| chr2:88315874..88316062 | 8.865 | RGPD1|RANBP2-like and GRIP domain containing 1 | -30660 (-) | KRCC1|lysine-rich coiled-coil 1 | 39291 (-) | |||||||||
| chr13:53191536..53191662 | 8.805 | CKAP2|cytoskeleton associated protein 2 | -156306 (+) | HNRNPA1L2|heterogeneous nuclear ribonucleoprotein A1-like 2 | 38 (+) | 18:0.202 |
||||||||
| chr16:2732411..2732535 | 8.799 | ERVK13-1|endogenous retrovirus group K13, member 1 | -9068 (-) | KCTD5|potassium channel tetramerisation domain containing 5 | 26 (+) | 40:0.823, -52:0.430, -57:0.224, -90:0.794 |
||||||||
| chr6:157744522..157744658 | 8.790 | TMEM242|transmembrane protein 242 | -6 (-) | TMEM242|transmembrane protein 242 | 627 (-) | 30:0.445 |
||||||||
| chr14:70826418..70826552 | 8.783 | COX16|COX16 cytochrome c oxidase assembly homolog (S. cerevisiae) | -65 (-) | SYNJ2BP-COX16|SYNJ2BP-COX16 readthrough SYNJ2BP|synaptojanin 2 binding protein | 57271 (-) | -75:0.506 |
-74:0.638 |
|||||||
| chr1:11968465..11968655 | 8.762 | NPPB|natriuretic peptide B | -49620 (-) | KIAA2013|KIAA2013 | 17913 (-) | |||||||||
| chr10:43278119..43278252 | 8.755 | BMS1|BMS1 homolog, ribosome assembly protein (yeast) | -232 (+) | BMS1|BMS1 homolog, ribosome assembly protein (yeast) | 65 (+) | 49:0.503 |
||||||||
| chr5:74062947..74063072 | 8.735 | GFM2|G elongation factor, mitochondrial 2 | -46 (-) | NSA2|NSA2 ribosome biogenesis homolog (S. cerevisiae) | 78 (+) | -18:0.331, -57:0.896 |
||||||||
| chr2:24299205..24299332 | 8.730 | SF3B14|splicing factor 3B, 14 kDa subunit | -97 (-) | SF3B14|splicing factor 3B, 14 kDa subunit | 53 (-) | 53:0.476, -55:0.660, 83:0.882 |
||||||||
| chr11:66189209..66189341 | 8.720 | NPAS4|neuronal PAS domain protein 4 | -714 (+) | MRPL11|mitochondrial ribosomal protein L11 | 17011 (-) | |||||||||
| chr1:163291561..163291692 | 8.716 | -65 (-) | NUF2|NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) | 150 (+) | -48:0.885, -115:0.496 |
|||||||||
| chr5:68855907..68856047 | 8.713 | OCLN|occludin | -55940 (+) | GTF2H2B|general transcription factor IIH, polypeptide 2B (pseudogene) GTF2H2C|general transcription factor IIH, polypeptide 2C GTF2H2D|general transcription factor IIH, polypeptide 2D | 64 (+) | 19:0.744, 64:0.371 |
-39:0.272 |
|||||||
| chr15:96289163..96289245 | 8.701 | LOC400456|uncharacterized LOC400456 | -418885 (-) | NR2F2|nuclear receptor subfamily 2, group F, member 2 | 579957 (+) | |||||||||
| chr6:131949358..131949506 | 8.695 | MED23|mediator complex subunit 23 | -145 (-) | ENPP3|ectonucleotide pyrophosphatase/phosphodiesterase 3 | 9050 (+) | -118:0.508 |
115:0.947 |
-71:0.550 |
||||||
| chr4:4543759..4543877 | 8.694 | STX18|syntaxin 18 | -57 (-) | LOC100507266|uncharacterized LOC100507266 | 66 (+) | |||||||||
| chr20:57226180..57226314 | 8.683 | APCDD1L|adenomatosis polyposis coli down-regulated 1-like | -136349 (-) | STX16-NPEPL1|STX16-NPEPL1 readthrough (non-protein coding) STX16|syntaxin 16 | 141 (+) | 21:0.663 |
-98:0.529 |
-91:0.273 |
||||||
| chr22:19419942..19420080 | 8.682 | MRPL40|mitochondrial ribosomal protein L40 | -629 (+) | MRPL40|mitochondrial ribosomal protein L40 | 36 (+) | 50:0.774 |
9:0.589 |
|||||||
| chr1:148604631..148604821 | 8.651 | NBPF15|neuroblastoma breakpoint family, member 15 NBPF16|neuroblastoma breakpoint family, member 16 | -27310 (+) | PPIAL4E|peptidylprolyl isomerase A (cyclophilin A)-like 4E | 39284 (+) | |||||||||
| chr3:53381602..53381745 | 8.644 | DCP1A|DCP1 decapping enzyme homolog A (S. cerevisiae) | -80 (-) | CACNA1D|calcium channel, voltage-dependent, L type, alpha 1D subunit | 146986 (+) | -28:0.555 |
||||||||
| chr1:45196424..45196615 | 8.634 | C1orf228|chromosome 1 open reading frame 228 | -56126 (+) | KIF2C|kinesin family member 2C | 8979 (+) | |||||||||
| chr1:147994416..147994606 | 8.626 | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | -39114 (-) | NBPF14|neuroblastoma breakpoint family, member 14 NBPF1|neuroblastoma breakpoint family, member 1 | 31343 (-) | |||||||||
| chr20:40322124..40322240 | 8.622 | CHD6|chromodomain helicase DNA binding protein 6 | -75121 (-) | PTPRT|protein tyrosine phosphatase, receptor type, T | 1496328 (-) | 50:0.496 |
||||||||
| chr1:148766672..148766862 | 8.602 | NBPF15|neuroblastoma breakpoint family, member 15 NBPF16|neuroblastoma breakpoint family, member 16 NBPF8|neuroblastoma breakpoint family, member 8 | -27266 (+) | PPIAL4D|peptidylprolyl isomerase A (cyclophilin A)-like 4D PPIAL4F|peptidylprolyl isomerase A (cyclophilin A)-like 4F | 39247 (+) | |||||||||
| chr11:68671280..68671406 | 8.601 | IGHMBP2|immunoglobulin mu binding protein 2 | -8 (+) | IGHMBP2|immunoglobulin mu binding protein 2 | 22 (+) | -45:0.505 |
||||||||
| chr13:31191951..31192064 | 8.594 | USPL1|ubiquitin specific peptidase like 1 | -44 (+) | ALOX5AP|arachidonate 5-lipoxygenase-activating protein | 95607 (+) | 11:0.300, 44:0.606, -51:0.912 |
||||||||
| chr13:46948594..46948687 | 8.582 | LCP1|lymphocyte cytosolic protein 1 (L-plastin) | -192248 (-) | KIAA0226L|KIAA0226-like | 12705 (-) | |||||||||
| chr17:61678149..61678279 | 8.582 | DCAF7|DDB1 and CUL4 associated factor 7 | -50202 (+) | TACO1|translational activator of mitochondrially encoded cytochrome c oxidase I | 18 (+) | 13:0.518 |
||||||||
| chr17:18966920..18967111 | 8.539 | GRAP|GRB2-related adaptor protein | -16729 (-) | GRAPL|GRB2-related adaptor protein-like | 63766 (+) | 75:0.240 |
||||||||
| chr5:70363504..70363634 | 8.527 | GTF2H2B|general transcription factor IIH, polypeptide 2B (pseudogene) GTF2H2C|general transcription factor IIH, polypeptide 2C GTF2H2|general transcription factor IIH, polypeptide 2, 44kDa | -115 (-) | BDP1|B double prime 1, subunit of RNA polymerase III transcription initiation factor IIIB | 387907 (+) | -14:0.744, -59:0.371 |
44:0.272 |
|||||||
| chr12:7052974..7053151 | 8.526 | -7606 (+) | C12orf57|chromosome 12 open reading frame 57 | 135 (+) | 100:0.797 |
|||||||||
| chr22:23241979..23242170 | 8.500 | IGLC1|immunoglobulin lambda constant 1 (Mcg marker) IGLL5|immunoglobulin lambda-like polypeptide 5 | -11991 (+) | GNAZ|guanine nucleotide binding protein (G protein), alpha z polypeptide | 170442 (+) | |||||||||
| chr1:78470069..78470212 | 8.497 | DNAJB4|DnaJ (Hsp40) homolog, subfamily B, member 4 | -25168 (+) | DNAJB4|DnaJ (Hsp40) homolog, subfamily B, member 4 | 474 (+) | |||||||||
| chr1:210547281..210547471 | 8.493 | HHAT|hedgehog acyltransferase | -44735 (+) | KCNH1|potassium voltage-gated channel, subfamily H (eag-related), member 1 | 760009 (-) | |||||||||
| chr1:17231638..17231829 | 8.483 | CROCCP2|ciliary rootlet coiled-coil, rootletin pseudogene 2 | -260556 (-) | CROCC|ciliary rootlet coiled-coil, rootletin | 16701 (+) | -51:0.555 |
||||||||
| chr5:69711064..69711195 | 8.465 | SMA4|glucuronidase, beta pseudogene | -14210 (+) | SMA4|glucuronidase, beta pseudogene | 170901 (-) | 14:0.744, 59:0.371 |
-44:0.272 |
|||||||
| chrUn_gl000219:99792..99983 | 8.465 | None | None (None) | None | None (None) | |||||||||
| chr1:149513847..149514030 | 8.463 | HIST2H2BF|histone cluster 2, H2bf | -114742 (-) | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | 39085 (+) | |||||||||
| chr17:8076474..8076664 | 8.458 | VAMP2|vesicle-associated membrane protein 2 (synaptobrevin 2) | -10277 (-) | TMEM107|transmembrane protein 107 | 3091 (-) | |||||||||
| chr9:108006607..108006740 | 8.438 | ABCA1|ATP-binding cassette, sub-family A (ABC1), member 1 | -316144 (-) | SLC44A1|solute carrier family 44, member 1 | 240 (+) | 22:0.209, -120:0.438 |
||||||||
| chr1:45187142..45187333 | 8.434 | C1orf228|chromosome 1 open reading frame 228 | -46844 (+) | KIF2C|kinesin family member 2C | 18261 (+) | |||||||||
| chr1:179851835..179851955 | 8.426 | TOR1AIP1|torsin A interacting protein 1 | -80 (+) | TOR1AIP1|torsin A interacting protein 1 | 21 (+) | 12:0.338, 35:0.730, 104:0.303 |
107:0.362 |
34:0.992 |
||||||
| chr14:24658116..24658229 | 8.422 | IPO4|importin 4 | -106 (-) | TM9SF1|transmembrane 9 superfamily member 1 | 6474 (-) | -15:0.546, -101:0.775 |
||||||||
| chr6:100016520..100016674 | 8.410 | CCNC|cyclin C | -140 (-) | CCNC|cyclin C | 6 (-) | 27:0.351 |
||||||||
| chr9:37800681..37800811 | 8.410 | EXOSC3|exosome component 3 | -15677 (-) | DCAF10|DDB1 and CUL4 associated factor 10 | 38 (+) | 40:0.896 |
||||||||
| chr16:89160174..89160328 | 8.402 | ACSF3|acyl-CoA synthetase family member 3 | -14 (+) | ACSF3|acyl-CoA synthetase family member 3 | 56 (+) | 21:0.522, 104:0.293 |
||||||||
| chr15:35261934..35262073 | 8.389 | AQR|aquarius homolog (mouse) | -76 (-) | ZNF770|zinc finger protein 770 | 18451 (-) | 20:0.809 |
89:0.239 |
|||||||
| chr17:29233229..29233370 | 8.382 | C17orf42|chromosome 17 open reading frame 42 | -48 (-) | ADAP2|ArfGAP with dual PH domains 2 | 15533 (+) | -40:0.736 |
||||||||
| chr15:65903442..65903573 | 8.374 | PTPLAD1|protein tyrosine phosphatase-like A domain containing 1 | -80584 (+) | C15orf44|chromosome 15 open reading frame 44 | 9 (-) | 7:0.334 |
||||||||
| chr3:194393170..194393310 | 8.367 | LSG1|large subunit GTPase 1 homolog (S. cerevisiae) | -35 (-) | FAM43A|family with sequence similarity 43, member A | 13383 (+) | 59:0.324 |
||||||||
| chr20:2451394..2451546 | 8.365 | SNRPB|small nuclear ribonucleoprotein polypeptides B and B1 | -38 (-) | SNRPB|small nuclear ribonucleoprotein polypeptides B and B1 | 3 (-) | 2:0.826, 29:0.299, -40:0.756, 120:0.367 |
||||||||
| chr1:212965126..212965263 | 8.363 | NSL1|NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) | -73 (-) | TATDN3|TatD DNase domain containing 3 | 24 (+) | -60:0.998 |
||||||||
| chr22:22337178..22337298 | 8.359 | TOP3B|topoisomerase (DNA) III beta | -52 (-) | 48127 (+) | -44:0.510 |
|||||||||
| chr2:197664394..197664532 | 8.356 | GTF3C3|general transcription factor IIIC, polypeptide 3, 102kDa | -50 (-) | C2orf66|chromosome 2 open reading frame 66 | 10474 (-) | -77:0.688, -96:0.279 |
14:0.442 |
|||||||
| chr10:14227689..14227879 | 8.344 | FRMD4A|FERM domain containing 4A | -177292 (-) | FRMD4A|FERM domain containing 4A | 145066 (-) | -46:0.234 |
||||||||
| chr3:185216746..185216862 | 8.339 | TMEM41A|transmembrane protein 41A | -105 (-) | LIPH|lipase, member H | 53524 (-) | |||||||||
| chr3:131221770..131221915 | 8.329 | MRPL3|mitochondrial ribosomal protein L3 | -88 (-) | CPNE4|copine IV | 531998 (-) | 1:0.912, 47:0.339 |
||||||||
| chr21:26948608..26948799 | 8.327 | MIR155HG|MIR155 host gene (non-protein coding) | -14231 (+) | MRPL39|mitochondrial ribosomal protein L39 | 31112 (-) | 62:0.214 |
||||||||
| chr5:133968507..133968614 | 8.320 | SAR1B|SAR1 homolog B (S. cerevisiae) | -63 (-) | SEC24A|SEC24 family, member A (S. cerevisiae) | 15871 (+) | -36:0.475 |
31:0.219 |
|||||||
| chr10:14227466..14227657 | 8.306 | FRMD4A|FERM domain containing 4A | -177069 (-) | FRMD4A|FERM domain containing 4A | 145289 (-) | 78:0.319 |
||||||||
| chr3:149470274..149470412 | 8.304 | COMMD2|COMM domain containing 2 | -73 (-) | ANKUB1|ankyrin repeat and ubiquitin domain containing 1 | 40266 (-) | -58:0.896 |
||||||||
| chr5:40835355..40835488 | 8.300 | RPL37|ribosomal protein L37 | -66 (-) | CARD6|caspase recruitment domain family, member 6 | 6021 (+) | -45:0.885, 59:0.239, -63:0.363, -87:0.751, -101:0.687 |
||||||||
| chr15:65588630..65588821 | 8.291 | PARP16|poly (ADP-ribose) polymerase family, member 16 | -9565 (-) | IGDCC3|immunoglobulin superfamily, DCC subclass, member 3 | 81626 (-) | |||||||||
| chr4:26585560..26585695 | 8.289 | CCKAR|cholecystokinin A receptor | -93642 (-) | TBC1D19|TBC1 domain family, member 19 | 53 (+) | 48:0.408 |
||||||||
| chr1:44679028..44679137 | 8.287 | KLF17|Kruppel-like factor 17 | -94533 (+) | DMAP1|DNA methyltransferase 1 associated protein 1 | 61 (+) | 12:0.912, 52:0.809 |
104:0.919 |
|||||||
| chr1:45285262..45285452 | 8.279 | BTBD19|BTB (POZ) domain containing 19 | -11147 (+) | PTCH2|patched 2 | 23280 (-) | |||||||||
| chr4:9927..10118 | 8.264 | None (None) | ZNF595|zinc finger protein 595 ZNF718|zinc finger protein 718 | 43202 (+) | ||||||||||
| chrUn_gl000219:100027..100173 | 8.264 | None | None (None) | None | None (None) | 25:0.247, -101:0.329, 102:0.338 |
||||||||
| chr11:57508760..57508899 | 8.261 | TMX2-CTNND1|TMX2-CTNND1 readthrough (non-protein coding) TMX2|thioredoxin-related transmembrane protein 2 | -28759 (+) | C11orf31|chromosome 11 open reading frame 31 | 23 (+) | -57:0.482 |
||||||||
| chr17:49230801..49230928 | 8.246 | SPAG9|sperm associated antigen 9 | -32709 (-) | NME1-NME2|NME1-NME2 readthrough NME1|non-metastatic cells 1, protein (NM23A) expressed in | 47 (+) | 3:0.553, -48:0.810, -73:0.275 |
||||||||
| chr7:129691167..129691294 | 8.245 | ZC3HC1|zinc finger, C3HC-type containing 1 | -9 (-) | KLHDC10|kelch domain containing 10 | 19130 (+) | 27:0.286 |
||||||||
| chr14:23039192..23039335 | 8.233 | -49914 (+) | DAD1|defender against cell death 1 | 18823 (-) | 19:0.332 |
|||||||||
| chr17:17991120..17991262 | 8.224 | C17orf39|chromosome 17 open reading frame 39 | -48384 (+) | DRG2|developmentally regulated GTP binding protein 2 | 77 (+) | 58:0.572, 95:0.220 |
||||||||
| chr13:53226925..53227066 | 8.219 | SUGT1|SGT1, suppressor of G2 allele of SKP1 (S. cerevisiae) | -53 (+) | LECT1|leukocyte cell derived chemotaxin 1 | 86869 (-) | 80:0.494 |
-118:0.387 |
70:0.232 |
||||||
| chr1:28975334..28975525 | 8.212 | TAF12|TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa | -5849 (-) | GMEB1|glucocorticoid modulatory element binding protein 1 | 19800 (+) | 56:0.267 |
||||||||
| chr11:65769628..65769757 | 8.202 | EIF1AD|eukaryotic translation initiation factor 1A domain containing | -62 (-) | BANF1|barrier to autointegration factor 1 | 127 (+) | 28:0.270, 52:0.296, -62:0.798 |
||||||||
| chr13:52768540..52768672 | 8.202 | THSD1P1|thrombospondin, type I, domain containing 1 pseudogene 1 | -47 (-) | THSD1|thrombospondin, type I, domain containing 1 | 211672 (-) | -9:0.588 |
||||||||
| chr15:75660926..75661049 | 8.187 | MAN2C1|mannosidase, alpha, class 2C, member 1 | -30 (-) | SIN3A|SIN3 transcription regulator homolog A (yeast) | 82915 (-) | 0:0.294 |
21:0.315 |
85:0.818 |
||||||
| chr17:57287297..57287406 | 8.180 | PRR11|proline rich 11 | -54231 (+) | SMG8|smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) | 46 (+) | -30:0.493, 35:0.364 |
-31:0.463 |
|||||||
| chr17:56756601..56756792 | 8.170 | C17orf47|chromosome 17 open reading frame 47 | -134989 (-) | TEX14|testis expressed 14 | 12699 (-) | |||||||||
| chr11:65274632..65274822 | 8.163 | -3217 (+) | SCYL1|SCY1-like 1 (S. cerevisiae) | 17810 (+) | ||||||||||
| chr5:102455840..102455980 | 8.158 | GIN1|gypsy retrotransposon integrase 1 | -97 (-) | PPIP5K2|diphosphoinositol pentakisphosphate kinase 2 | 71 (+) | 18:0.500, -76:0.768, -104:0.380 |
-52:0.216 |
|||||||
| chr8:33371036..33371224 | 8.158 | TTI2|TELO2 interacting protein 2 | -503 (-) | RNF122|ring finger protein 122 | 53533 (-) | |||||||||
| chr19:50879538..50879664 | 8.157 | NAPSA|napsin A aspartic peptidase | -10698 (-) | NR1H2|nuclear receptor subfamily 1, group H, member 2 | 126 (+) | 16:0.836, 34:0.301, -36:0.368, 75:0.836, 107:0.438 |
||||||||
| chr10:95462357..95462472 | 8.130 | FRA10AC1|fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 | -79 (-) | LGI1|leucine-rich, glioma inactivated 1 | 55232 (+) | 48:0.215, -66:0.912 |
||||||||
| chr1:999888..999971 | 8.121 | LOC100288175|uncharacterized LOC100288175 | -4817 (+) | RNF223|ring finger protein 223 | 9757 (-) | -7:0.622 |
||||||||
| chr15:41694669..41694792 | 8.119 | NDUFAF1|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 | -68 (-) | RTF1|Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | 14630 (+) | 19:0.351, 42:0.270 |
||||||||
| chr13:113862979..113863108 | 8.112 | PCID2|PCI domain containing 2 | -57 (-) | CUL4A|cullin 4A | 58 (+) | -31:0.896 |
||||||||
| chr21:26979781..26979925 | 8.102 | MRPL39|mitochondrial ribosomal protein L39 | -38 (-) | JAM2|junctional adhesion molecule 2 | 31800 (+) | -13:0.288 |
62:0.582 |
|||||||
| chr1:227922986..227923112 | 8.097 | JMJD4|jumonji domain containing 4 | -78 (-) | JMJD4|jumonji domain containing 4 | 50 (-) | -24:0.295, -36:0.715, -112:0.255 |
||||||||
| chr5:149780345..149780535 | 8.095 | TCOF1|Treacher Collins-Franceschetti syndrome 1 | -43116 (+) | CD74|CD74 molecule, major histocompatibility complex, class II invariant chain | 11863 (-) | |||||||||
| chr20:31989334..31989427 | 8.093 | CDK5RAP1|CDK5 regulatory subunit associated protein 1 | -41 (-) | SNTA1|syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | 42181 (-) | -7:0.495, -45:0.311 |
||||||||
| chr4:25314309..25314440 | 8.090 | PI4K2B|phosphatidylinositol 4-kinase type 2 beta | -78656 (+) | ZCCHC4|zinc finger, CCHC domain containing 4 | 73 (+) | -31:0.378, 56:0.228 |
||||||||
| chr19:11039373..11039510 | 8.078 | YIPF2|Yip1 domain family, member 2 | -202 (-) | C19orf52|chromosome 19 open reading frame 52 | 16 (+) | -40:0.912 |
||||||||
| chrX:12993298..12993488 | 8.069 | TMSB4X|thymosin beta 4, X-linked | -153 (+) | TMSB4X|thymosin beta 4, X-linked | 950 (+) | |||||||||
| chr1:1310758..1310929 | 8.065 | AURKAIP1|aurora kinase A interacting protein 1 | -270 (-) | AURKAIP1|aurora kinase A interacting protein 1 | 20 (-) | 7:0.342, 46:0.501, 68:0.304, 89:0.546, 112:0.535 |
-42:0.224 |
|||||||
| chr1:206785771..206785884 | 8.064 | EIF2D|eukaryotic translation initiation factor 2D | -72 (-) | EIF2D|eukaryotic translation initiation factor 2D | 56 (-) | -62:0.503 |
||||||||
| chr2:55496339..55496466 | 8.063 | MTIF2|mitochondrial translational initiation factor 2 | -37 (-) | CCDC88A|coiled-coil domain containing 88A | 63384 (-) | 9:0.648, -40:0.422 |
||||||||
| chr14:106319928..106320119 | 8.061 | IGHM|immunoglobulin heavy constant mu | -82319 (-) | 1658 (-) | ||||||||||
| chr16:2653279..2653422 | 8.045 | PDPK1|3-phosphoinositide dependent protein kinase-1 | -65243 (+) | LOC652276|potassium channel tetramerisation domain containing 5 pseudogene | 67 (+) | -3:0.551, -41:0.782, 85:0.687 |
-51:0.419 |
|||||||
| chr16:740391..740500 | 8.043 | WDR24|WD repeat domain 24 | -43 (-) | FBXL16|F-box and leucine-rich repeat protein 16 | 15370 (-) | 9:0.770, -22:0.896, 46:0.319, -51:0.606 |
||||||||
| chr17:42092311..42092456 | 8.037 | TMEM101|transmembrane protein 101 | -17 (-) | LSM12|LSM12 homolog (S. cerevisiae) | 51583 (-) | 9:0.628, 41:0.308 |
||||||||
| chr1:150209425..150209536 | 8.034 | ANP32E|acidic (leucine-rich) nuclear phosphoprotein 32 family, member E | -988 (-) | CA14|carbonic anhydrase XIV | 20753 (+) | -53:0.390 |
||||||||
| chr4:164415676..164415830 | 8.034 | C4orf43|chromosome 4 open reading frame 43 | -35 (+) | C4orf43|chromosome 4 open reading frame 43 | 63 (+) | 63:0.218, -65:0.221 |
||||||||
| chr4:78783795..78783986 | 8.017 | CNOT6L|CCR4-NOT transcription complex, subunit 6-like | -43346 (-) | MRPL1|mitochondrial ribosomal protein L1 | 24 (+) | -96:0.215 |
-3:0.313 |
|||||||
| chr3:155572258..155572388 | 8.014 | SLC33A1|solute carrier family 33 (acetyl-CoA transporter), member 1 | -117 (-) | GMPS|guanine monphosphate synthetase | 15978 (+) | -80:0.768, -98:0.792 |
||||||||
| chr19:1383782..1383904 | 7.989 | MUM1|melanoma associated antigen (mutated) 1 | -28842 (+) | NDUFS7|NADH dehydrogenase (ubiquinone) Fe-S protein 7, 20kDa (NADH-coenzyme Q reductase) | 43 (+) | -19:0.219 |
||||||||
| chr4:83955984..83956143 | 7.985 | LIN54|lin-54 homolog (C. elegans) | -22035 (-) | COPS4|COP9 constitutive photomorphogenic homolog subunit 4 (Arabidopsis) | 261 (+) | |||||||||
| chr8:95731987..95732098 | 7.958 | ESRP1|epithelial splicing regulatory protein 1 | -78598 (+) | DPY19L4|dpy-19-like 4 (C. elegans) | 98 (+) | -57:0.300, 66:0.770 |
||||||||
| chr20:1099108..1099252 | 7.952 | PSMF1|proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | -5220 (+) | PSMF1|proteasome (prosome, macropain) inhibitor subunit 1 (PI31) | 173 (+) | -51:0.606, 62:0.797 |
||||||||
| chr9:114393400..114393523 | 7.951 | PTGR1|prostaglandin reductase 1 | -31344 (-) | DNAJC25-GNG10|DNAJC25-GNG10 readthrough DNAJC25|DnaJ (Hsp40) homolog, subfamily C , member 25 | 154 (+) | 32:0.684, 83:0.291 |
-3:0.865 |
|||||||
| chr9:140473318..140473484 | 7.944 | WDR85|WD repeat domain 85 | -29 (-) | ZMYND19|zinc finger, MYND-type containing 19 | 11531 (-) | -19:0.293 |
58:0.541 |
|||||||
| chr1:150459668..150459813 | 7.942 | RPRD2|regulation of nuclear pre-mRNA domain containing 2 | -122518 (+) | TARS2|threonyl-tRNA synthetase 2, mitochondrial (putative) | 152 (+) | -16:0.457, 45:0.249 |
52:0.215 |
15:0.301 |
||||||
| chr1:143673162..143673283 | 7.937 | -205597 (-) | FLJ39739|uncharacterized FLJ39739 LOC100286793|uncharacterized LOC100286793 | 71183 (-) | ||||||||||
| chr13:41345297..41345429 | 7.930 | MRPS31|mitochondrial ribosomal protein S31 | -31 (-) | SLC25A15|solute carrier family 25 (mitochondrial carrier; ornithine transporter) member 15 | 18226 (+) | -7:0.705 |
||||||||
| chr21:46359832..46359937 | 7.925 | C21orf67|chromosome 21 open reading frame 67 | -121 (-) | FAM207A|family with sequence similarity 207, member A | 41 (+) | -23:0.798, -33:0.774, 47:0.627 |
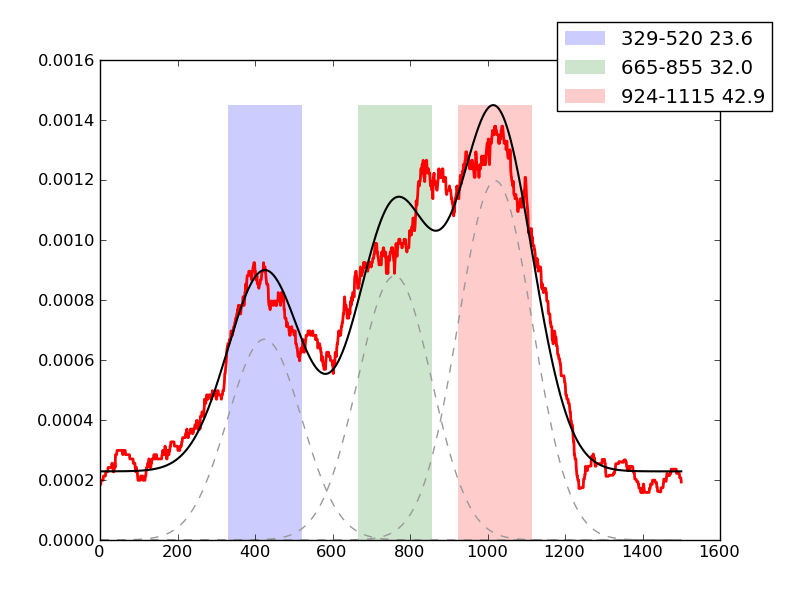
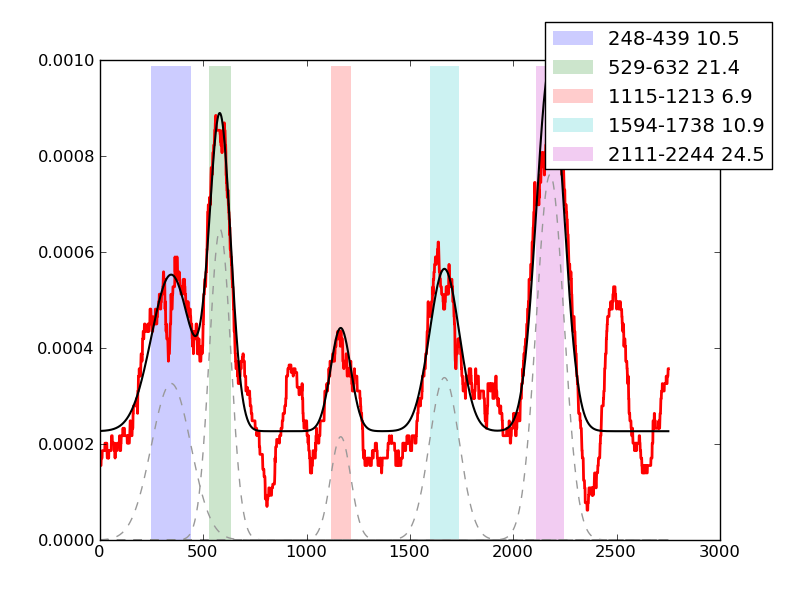
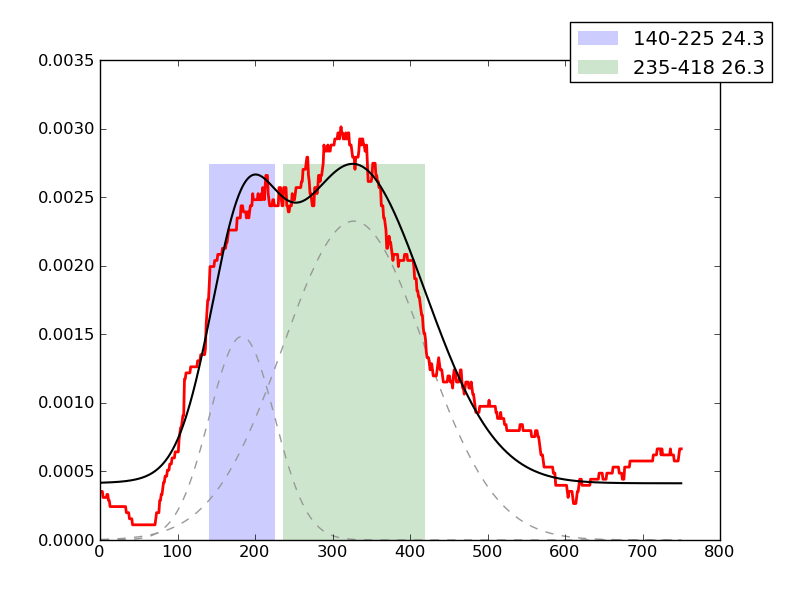
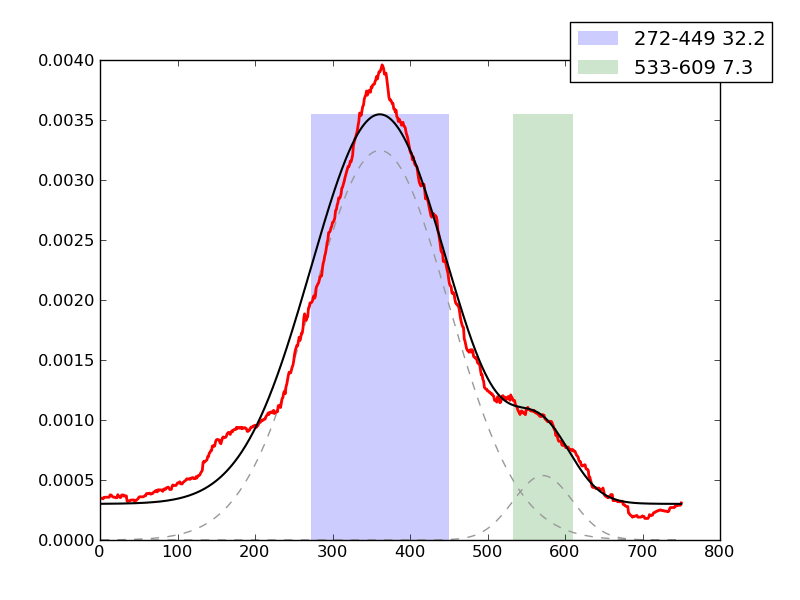
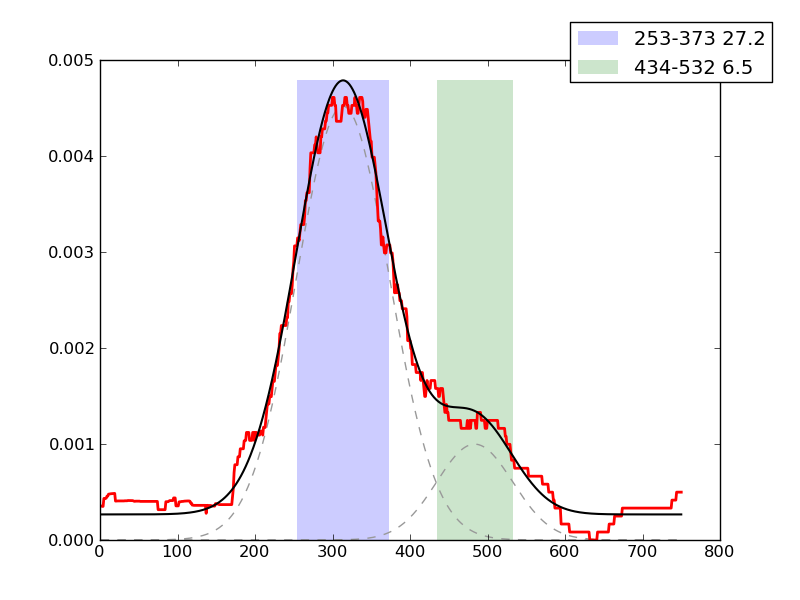
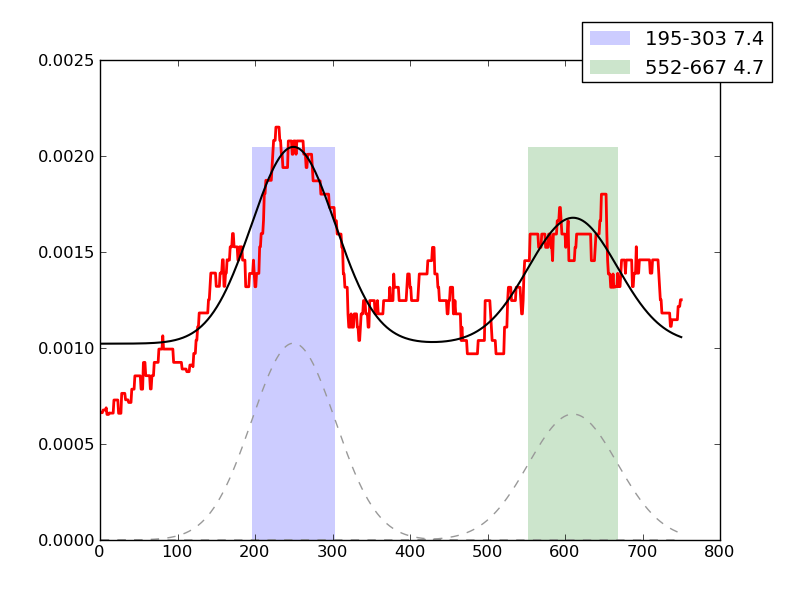
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| HCMC.ELF1_f1.wm | 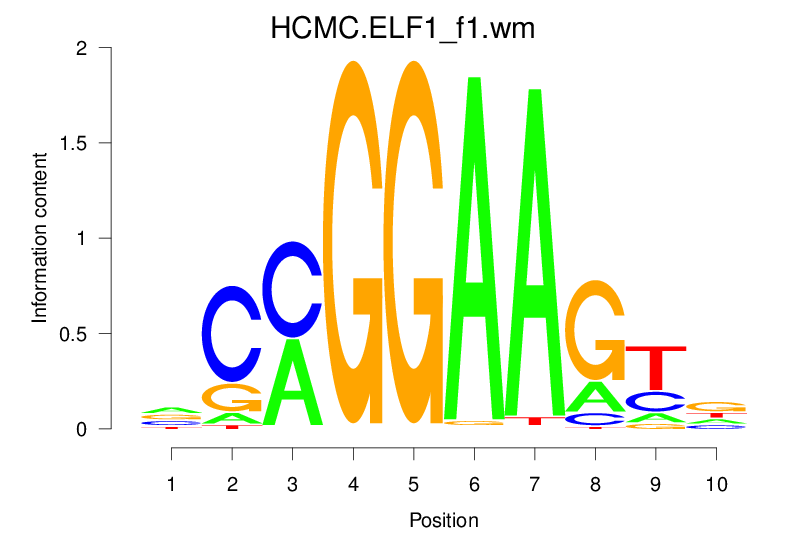 |
1.76 (281.449) | 1.76 (281.449) | 0.3921 | 0.12 | 1.4332 | 9381/19318 |
| HCMC.IRF8_si.wm | 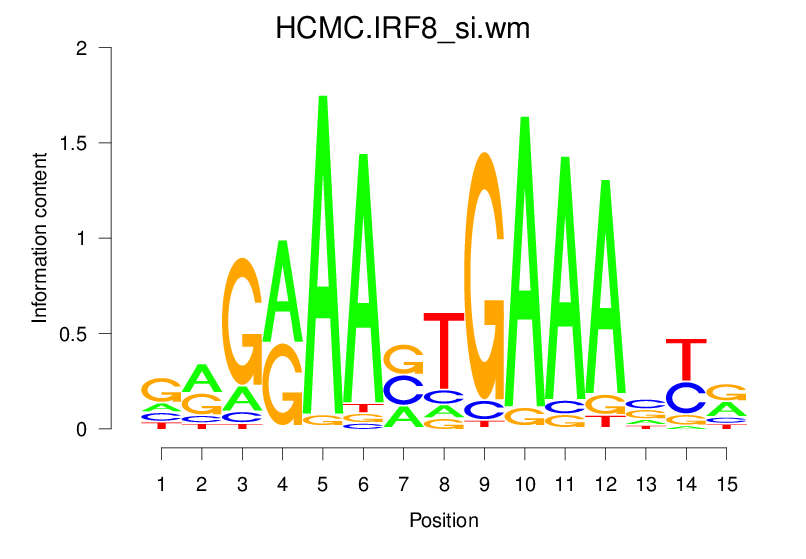 |
2.065 (361.044) | 1.213 (96.319) | 0.2398 | -0.0063 | 1.3902 | 2213/19318 |
| ZNF143.p2 | 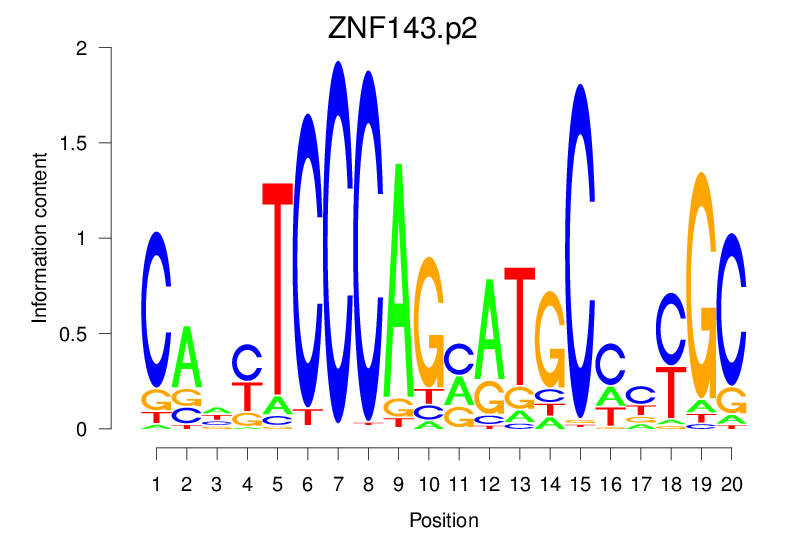 |
2.296 (413.814) | 1.101 (47.801) | 0.1787 | 0.0528 | 1.8131 | 870/19318 |
| denovo_WM_17 | 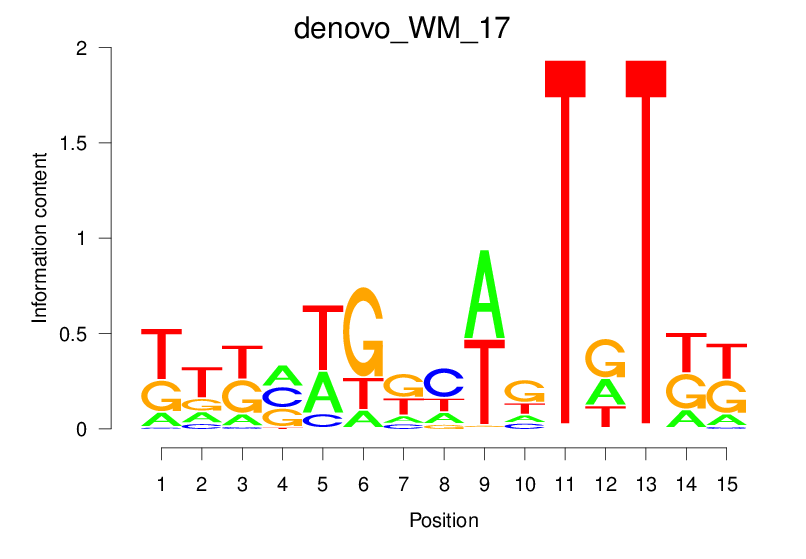 |
2.574 (470.775) | 1.079 (38.009) | 0.1628 | 0.0261 | 1.1385 | 696/19318 |
| HCMC.CTCF_f2.wm | 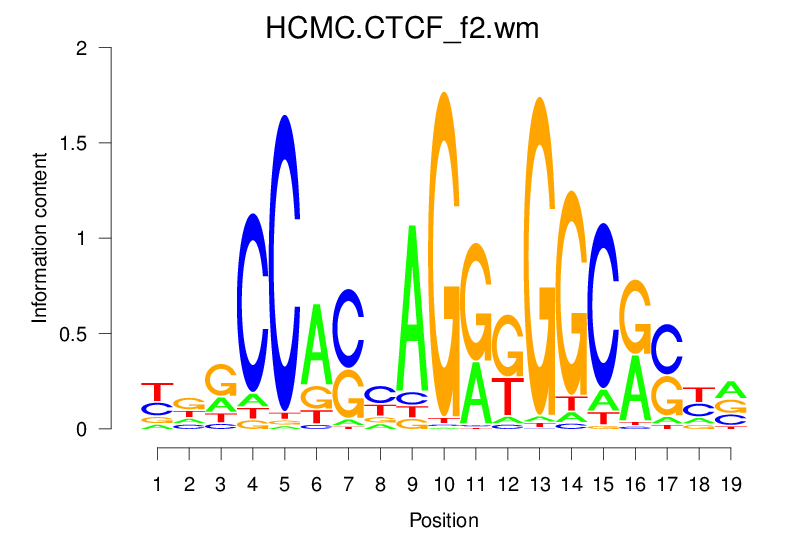 |
2.756 (504.833) | 1.136 (63.708) | 0.2121 | 0.0079 | 1.6575 | 3715/19318 |
| denovo_WM_21 | 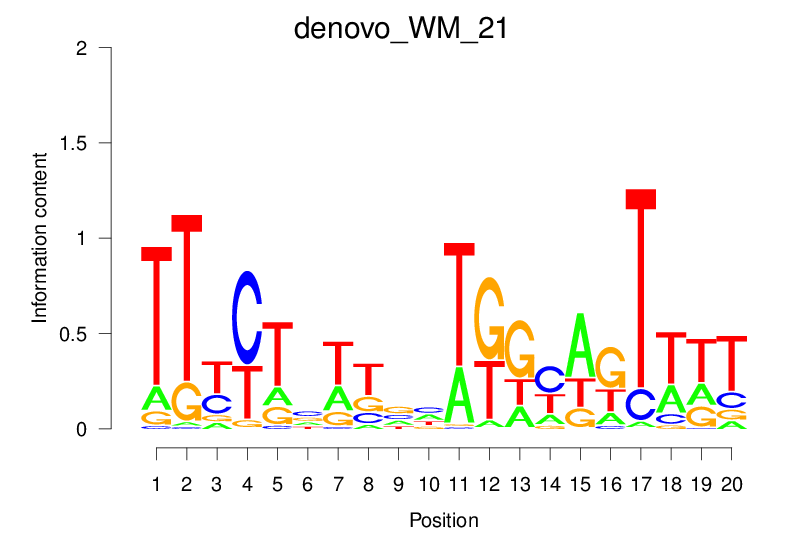 |
2.888 (528.075) | 1.087 (41.401) | 0.1797 | 0.0193 | 0.8191 | 871/19318 |
| HCMC.SPI1_si.wm | 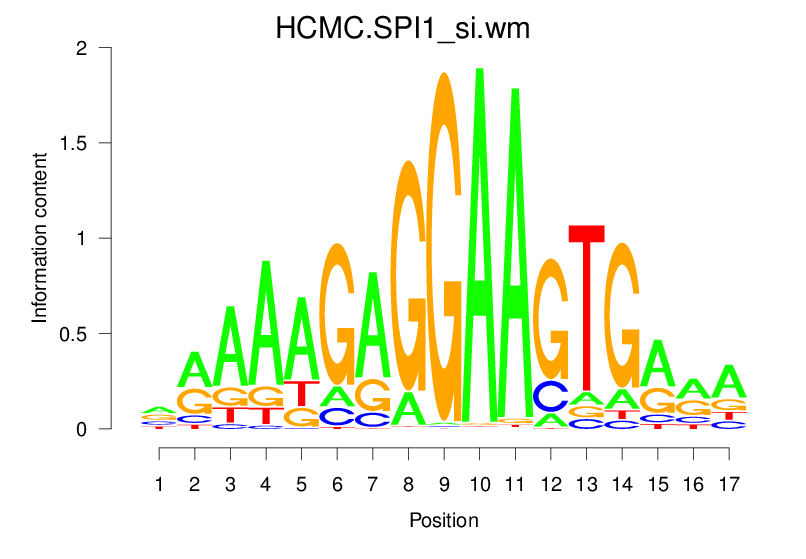 |
3.011 (549.002) | 1.253 (112.137) | 0.2631 | -0.0111 | 1.4773 | 1829/19318 |
| JASPAR.Myf.wm | 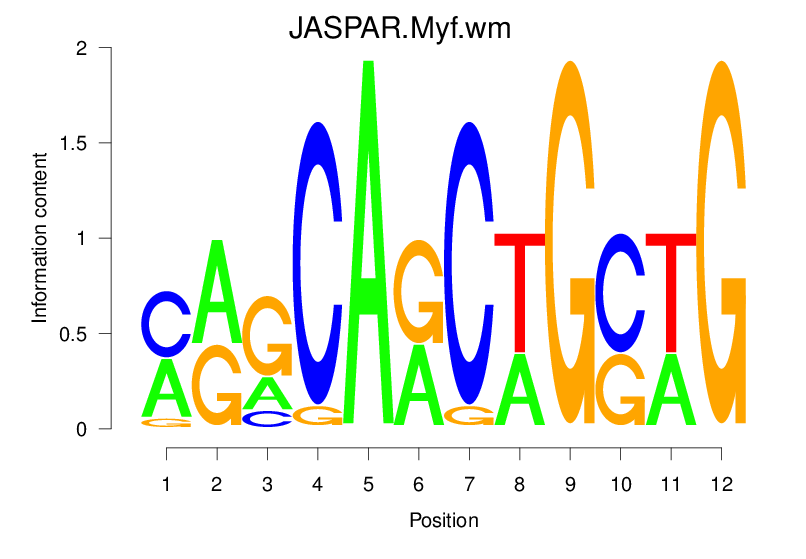 |
3.122 (566.951) | 1.067 (32.387) | 0.1827 | -0.0098 | 1.2066 | 3830/19318 |
| HCMC.FOXM1_f1.wm | 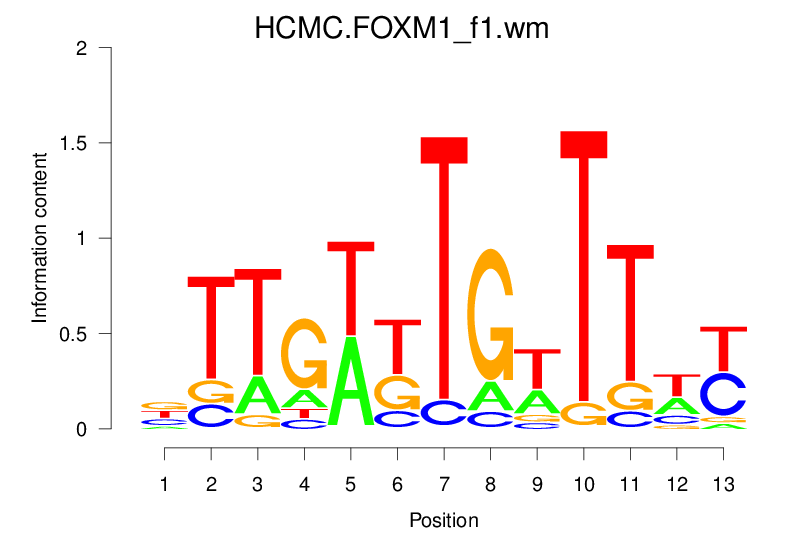 |
3.207 (580.41) | 1.034 (16.755) | 0.1466 | -0.0201 | 0.8306 | 69/19318 |
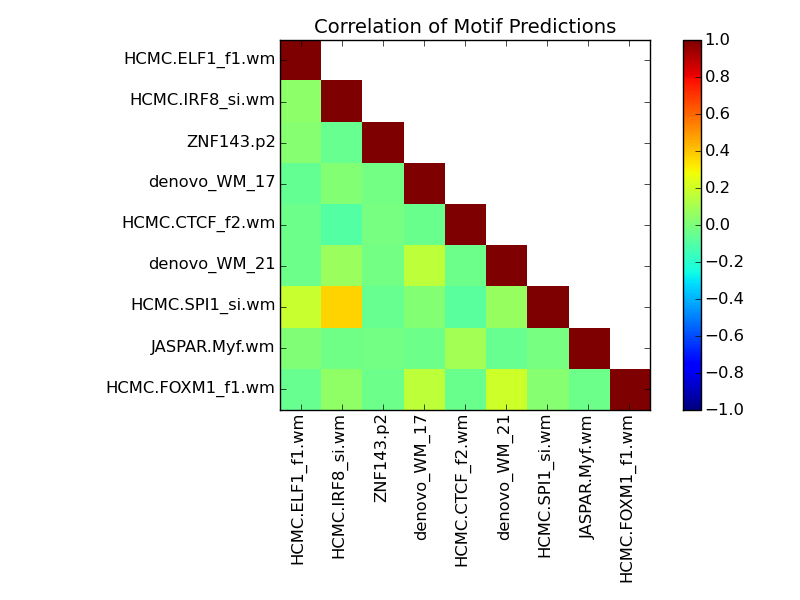
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| HCMC.ELF1_f1.wm |  |
1.76 (281.449) | 0.3921 | 0.12 | 1.4332 | 9381/19318 |
| HCMC.ETV4_f1.wm | 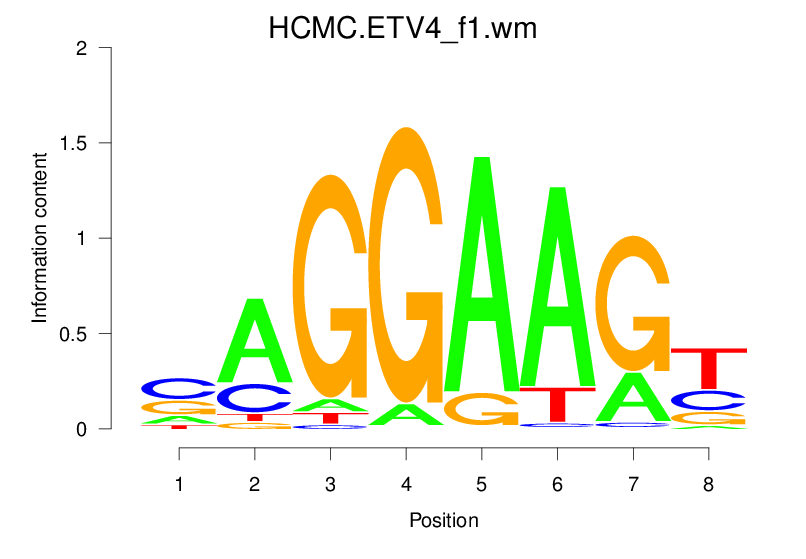 |
1.729 (272.624) | 0.4179 | 0.0645 | 1.1907 | 18436/19318 |
| HCMC.GABP1+GABP2_f1.wm | 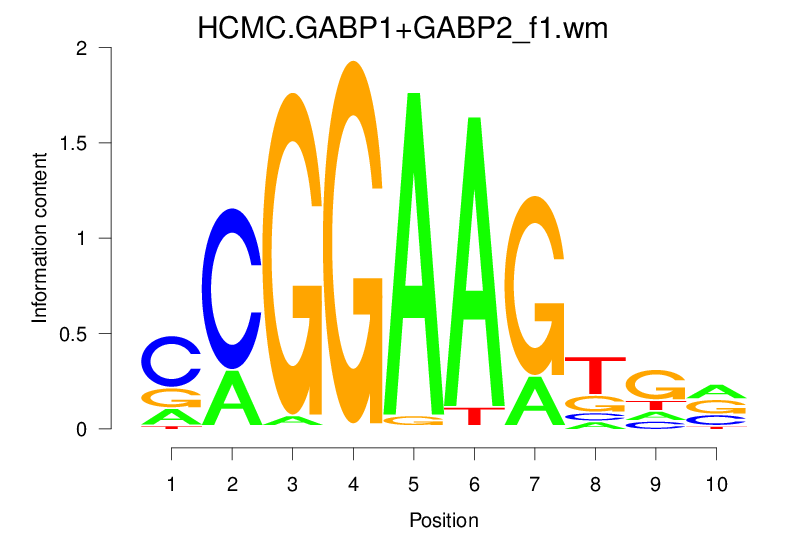 |
1.679 (258.053) | 0.3703 | 0.1238 | 1.3765 | 10867/19318 |
| JASPAR.SPI1.wm | 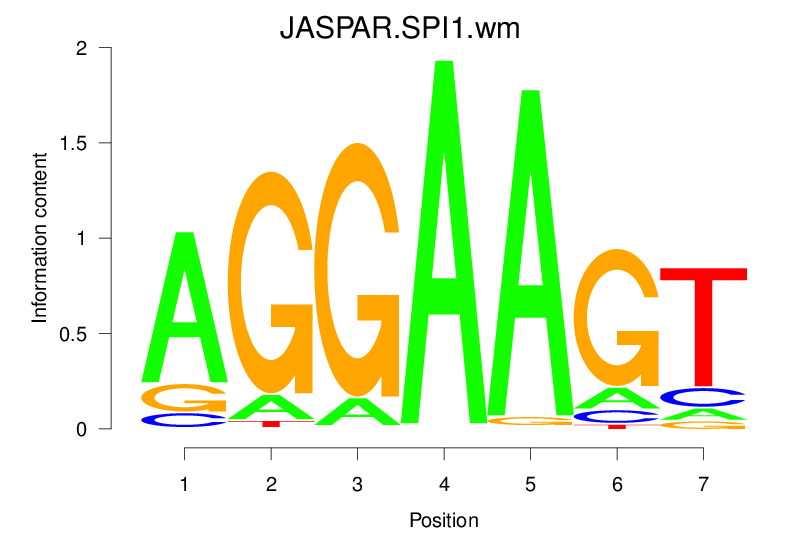 |
1.638 (245.834) | 0.3048 | 0.0013 | 1.1925 | 10904/19318 |
| denovo_WM_5 | 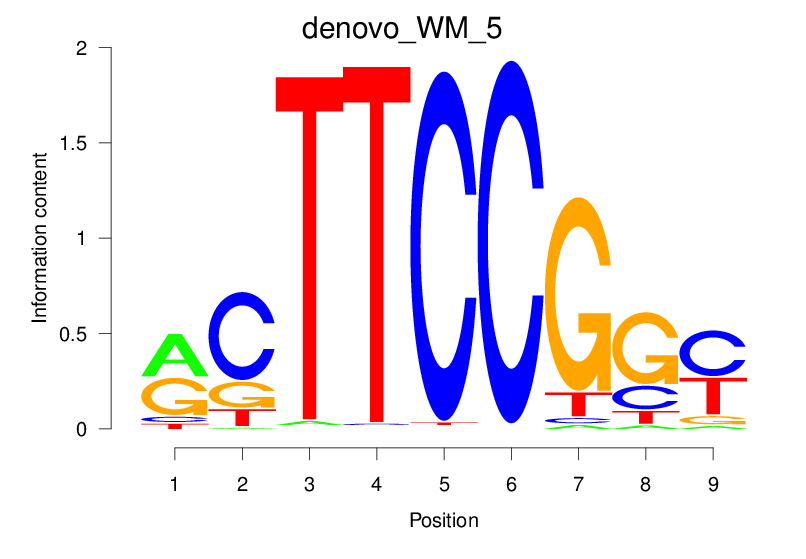 |
1.617 (239.477) | 0.3593 | 0.1644 | 1.6543 | 5806/19318 |
| HCMC.ELK1_f1.wm | 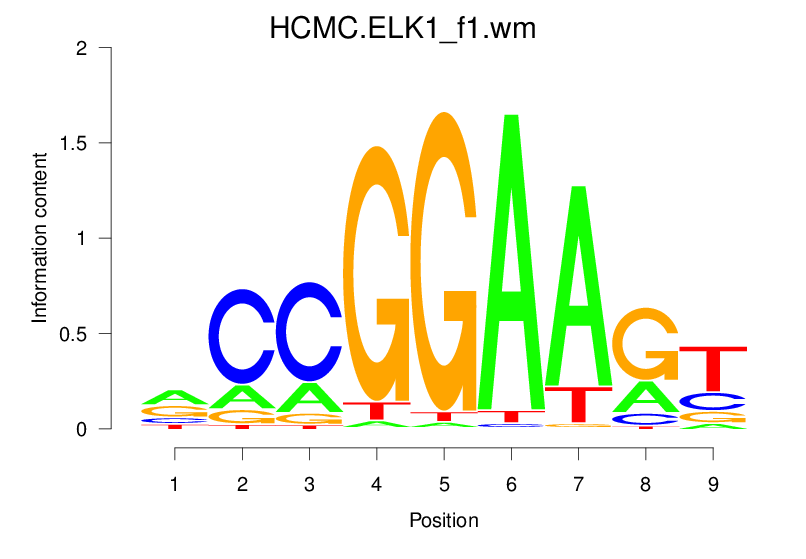 |
1.616 (239.007) | 0.3720 | 0.1515 | 1.4135 | 10885/19318 |
| HCMC.ETS1_si.wm | 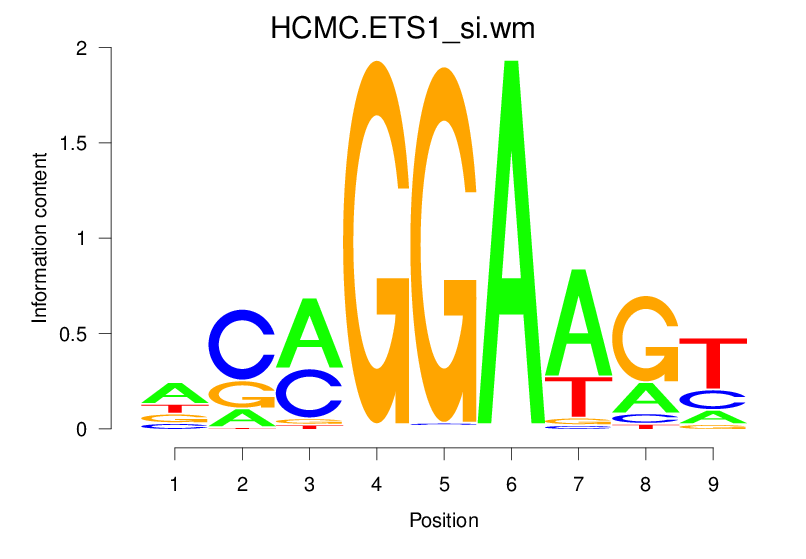 |
1.613 (238.177) | 0.3673 | 0.0887 | 1.3254 | 10917/19318 |
| HCMC.GABPA_f1.wm | 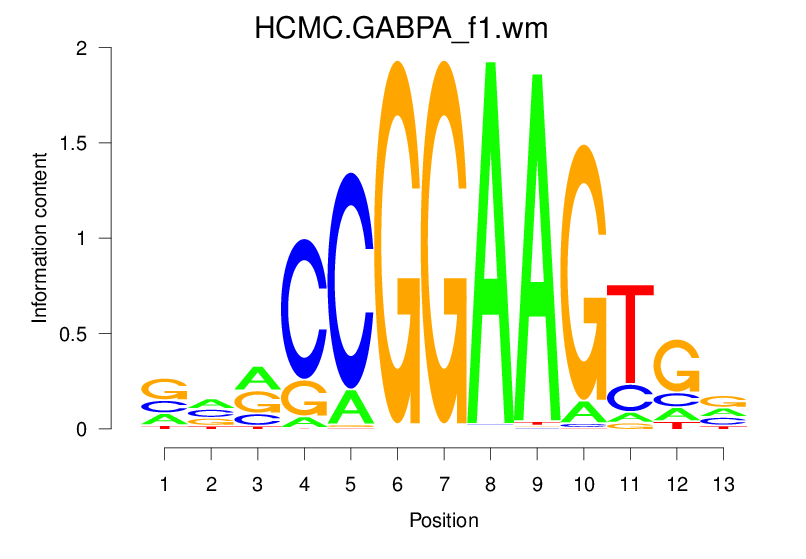 |
1.611 (237.334) | 0.3476 | 0.1343 | 1.8067 | 3376/19318 |
| denovo_WM_23 | 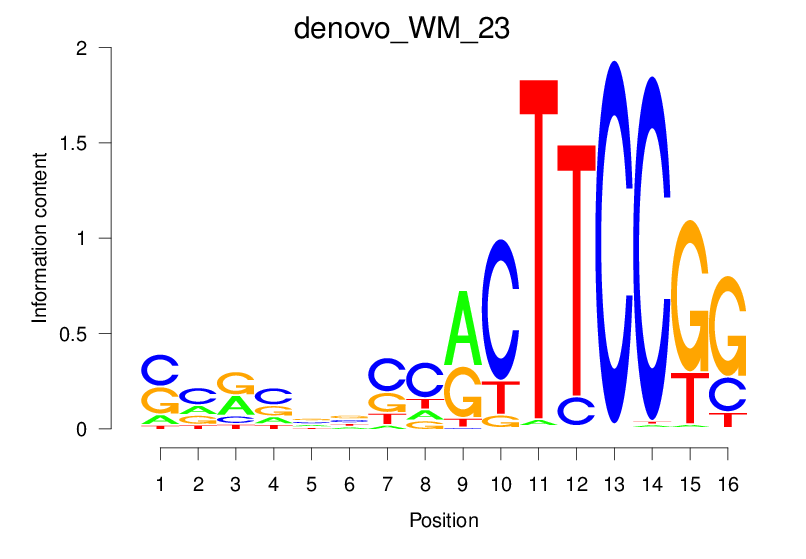 |
1.601 (234.32) | 0.3532 | 0.1293 | 1.5143 | 6335/19318 |
| HCMC.ETS2_f1.wm | 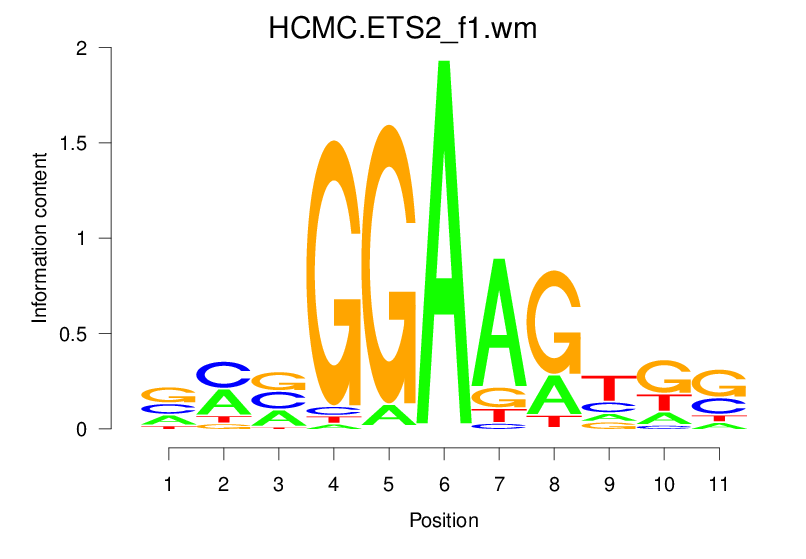 |
1.595 (232.365) | 0.3815 | 0.0775 | 1.1818 | 18157/19318 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis