CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.933 | 93rd percentile | |
Fraction of mapped reads for the background samples: |
0.714 | 37th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.227 | 61st percentile | |
Error in fit of enrichment distribution: |
0.509 | 45th percentile | |
Peak statistics |
|||
Number of peaks: |
37435 | 82nd percentile | |
Fraction of reads mapped to peaks: |
0.069 | 84th percentile | |
Motif enrichment statistics |
|||
Enrichment of top motif: |
2.95 | 36th percentile | |
Enrichment of complimentary motif set: |
13.33 | 62nd percentile | |
Quality Control, Mapping, Fragment Size Estimation:
CHD2 Replicate 1
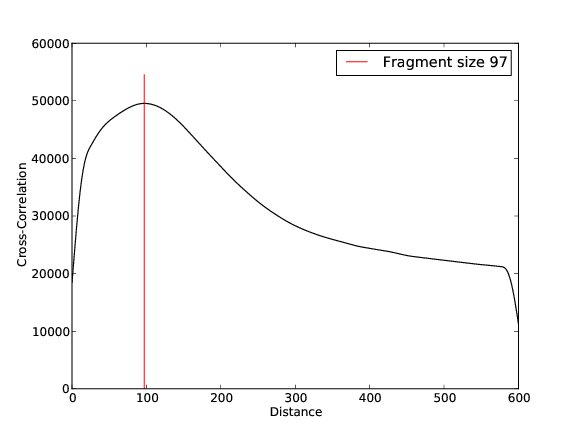
- 14,263,018 input reads (in FASTQ format).
- 14,196,800 reads after removal of low quality reads.
- 14,149,758 reads after removal of adapters and low complexity reads.
- 13,237,522 reads after mapping.
- Report PDF
CHD2 Replicate 2
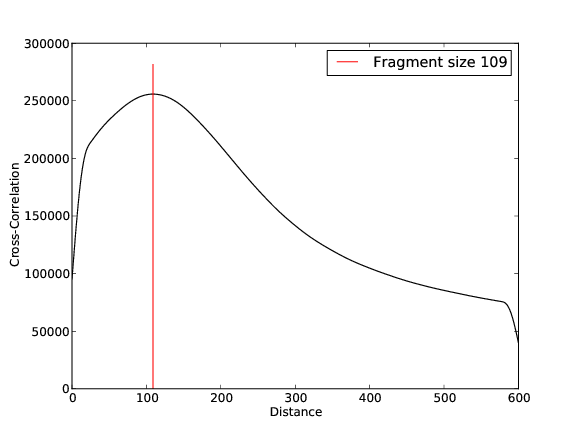
- 21,944,812 input reads (in FASTQ format).
- 21,756,670 reads after removal of low quality reads.
- 21,706,161 reads after removal of adapters and low complexity reads.
- 20,540,046 reads after mapping.
- Report PDF
BG Replicate 1
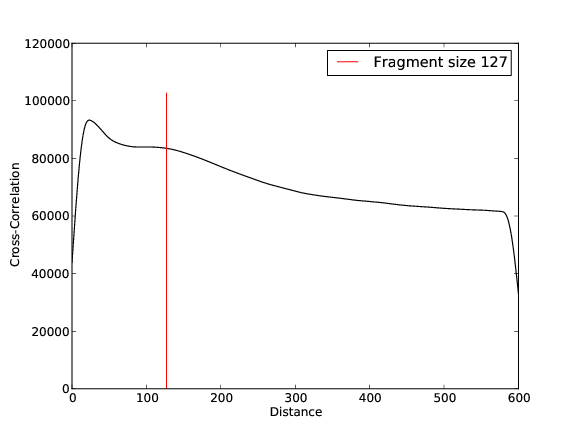
- 55,865,436 input reads (in FASTQ format).
- 44,385,915 reads after removal of low quality reads.
- 44,200,443 reads after removal of adapters and low complexity reads.
- 39,862,320 reads after mapping.
- Report PDF
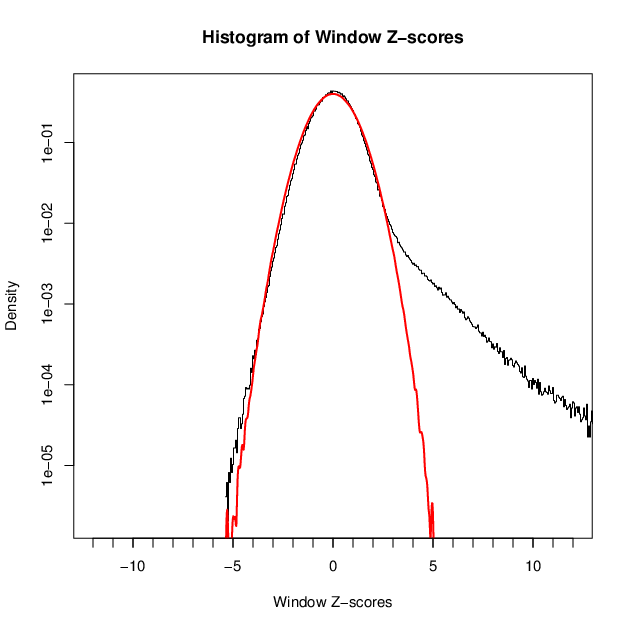
Peak Calling:
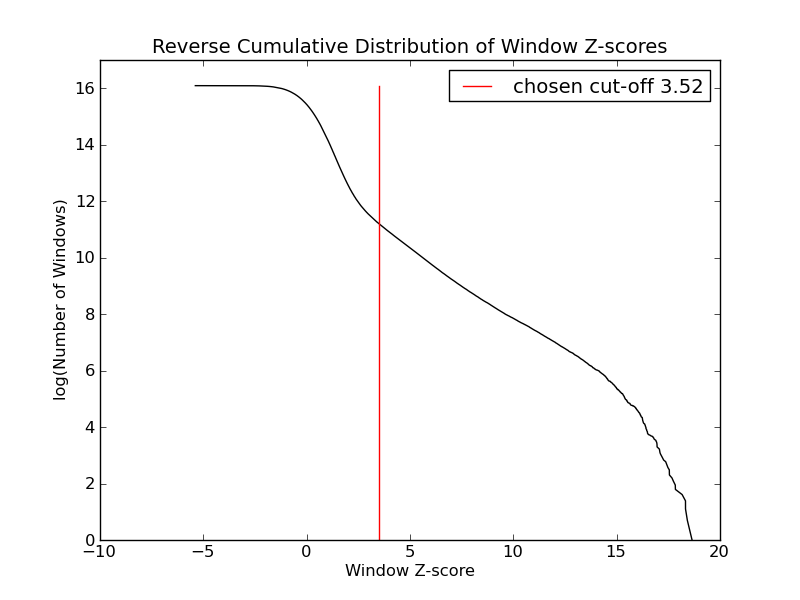
- 72172 windows are significantly enriched at FDR=10% (z-value cut-off: 3.52)
- 72172 windows merged to 26971 regions.
- 37435 significant peaks fitted within 26971 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | denovo_WM_21 | HCMC.NFYA_f1.wm | denovo_WM_17 | HTSELEX.JDP2.bZIP.full.dimeric.wm2 | HTSELEX.ETV6.ETS.full.dimeric.wm1 | HOMER.RLR1?.SacCer-Promoters |
|---|---|---|---|---|---|---|---|---|---|---|---|
| chr8:144911500..144911617 | 18.828 | PUF60|poly-U binding splicing factor 60KDa | -8 (-) | NRBP2|nuclear receptor binding protein 2 | 11601 (-) | 4:0.941, -24:0.924 |
|||||
| chr10:135186876..135186989 | 18.433 | ECHS1|enoyl CoA hydratase, short chain, 1, mitochondrial | -46 (-) | PAOX|polyamine oxidase (exo-N4-amino) | 5825 (+) | -9:0.217, 9:0.606, 12:0.351 |
|||||
| chr12:123237280..123237402 | 18.197 | HCAR1|hydroxycarboxylic acid receptor 1 | -22336 (-) | DENR|density-regulated protein | 50 (+) | 13:0.792, -14:0.923, 37:0.896 |
|||||
| chr9:6413098..6413210 | 18.050 | UHRF2|ubiquitin-like with PHD and ring finger domains 2 | -10 (+) | UHRF2|ubiquitin-like with PHD and ring finger domains 2 | 43 (+) | 0:0.497, -3:0.497 |
|||||
| chr12:133263910..133264041 | 17.773 | POLE|polymerase (DNA directed), epsilon | -35 (-) | POLE|polymerase (DNA directed), epsilon | 46 (-) | -10:0.305, -13:0.434 |
|||||
| chr19:4402454..4402583 | 17.750 | SH3GL1|SH3-domain GRB2-like 1 | -1965 (-) | CHAF1A|chromatin assembly factor 1, subunit A (p150) | 198 (+) | 15:0.572, 18:0.363 |
-66:0.865 |
||||
| chr19:4124123..4124230 | 17.661 | MAP2K2|mitogen-activated protein kinase kinase 2 | -43 (-) | CREB3L3|cAMP responsive element binding protein 3-like 3 | 29491 (+) | -8:0.837 |
|||||
| chr6:35995385..35995553 | 17.647 | SLC26A8|solute carrier family 26, member 8 | -3129 (-) | MAPK14|mitogen-activated protein kinase 14 | 4 (+) | -36:0.974, 65:0.276, 68:0.714 |
|||||
| chr15:75287740..75287843 | 17.575 | RPP25|ribonuclease P/MRP 25kDa subunit | -38017 (-) | SCAMP5|secretory carrier membrane protein 5 | 116 (+) | -7:0.788, 32:0.830 |
|||||
| chr10:124768306..124768421 | 17.454 | IKZF5|IKAROS family zinc finger 5 (Pegasus) | -92 (-) | ACADSB|acyl-CoA dehydrogenase, short/branched chain | 85 (+) | -7:0.416, -10:0.227 |
|||||
| chr13:113951441..113951561 | 17.354 | LAMP1|lysosomal-associated membrane protein 1 | -14 (+) | LAMP1|lysosomal-associated membrane protein 1 | 45 (+) | 8:0.460, 11:0.530 |
|||||
| chr4:2965104..2965220 | 17.326 | NOP14|NOP14 nucleolar protein homolog (yeast) | -39 (-) | NOP14|NOP14 nucleolar protein homolog (yeast) | 32 (-) | -12:0.838 |
|||||
| chr11:13484799..13484916 | 17.322 | BTBD10|BTB (POZ) domain containing 10 | -66 (-) | PTH|parathyroid hormone | 32688 (-) | 6:0.489, 9:0.489, -33:0.236 |
|||||
| chr1:6614552..6614663 | 17.297 | NOL9|nucleolar protein 9 | -44 (-) | NOL9|nucleolar protein 9 | 13 (-) | -7:0.474, -10:0.520 |
|||||
| chr7:158497455..158497587 | 17.245 | NCAPG2|non-SMC condensin II complex, subunit G2 | -44 (-) | ESYT2|extended synaptotagmin-like protein 2 | 113849 (-) | 2:0.858 |
|||||
| chr12:129308525..129308628 | 17.169 | SLC15A4|solute carrier family 15, member 4 | -133 (-) | GLT1D1|glycosyltransferase 1 domain containing 1 | 29408 (+) | -14:0.938 |
|||||
| chr17:40714048..40714160 | 17.125 | HSD17B1|hydroxysteroid (17-beta) dehydrogenase 1 | -7417 (-) | COASY|CoA synthase | 13 (+) | -15:0.927 |
|||||
| chr13:21750697..21750811 | 17.096 | SKA3|spindle and kinetochore associated complex subunit 3 | -66 (-) | MRP63|mitochondrial ribosomal protein 63 | 46 (+) | -7:0.853, 29:0.949 |
|||||
| chr2:120517221..120517348 | 17.013 | LOC100125918|uncharacterized LOC100125918 TMEM177|transmembrane protein 177 | -80503 (+) | PTPN4|protein tyrosine phosphatase, non-receptor type 4 (megakaryocyte) | 116 (+) | 1:0.903 |
|||||
| chr16:90088960..90089072 | 16.985 | DBNDD1|dysbindin (dystrobrevin binding protein 1) domain containing 1 | -2703 (-) | GAS8|growth arrest-specific 8 | 45 (+) | 0:0.403, 2:0.531 |
|||||
| chr12:49351287..49351392 | 16.936 | ARF3|ADP-ribosylation factor 3 | -35 (-) | WNT10B|wingless-type MMTV integration site family, member 10B | 14259 (-) | 11:0.712, 14:0.283, -18:0.631 |
|||||
| chr7:75677304..75677417 | 16.936 | STYXL1|serine/threonine/tyrosine interacting-like 1 | -25 (-) | MDH2|malate dehydrogenase 2, NAD (mitochondrial) | 18 (+) | 14:0.301, 17:0.642, -26:0.400, -29:0.217 |
|||||
| chr14:102414450..102414578 | 16.905 | PPP2R5C|protein phosphatase 2, regulatory subunit B', gamma | -138205 (+) | DYNC1H1|dynein, cytoplasmic 1, heavy chain 1 | 16314 (+) | -7:0.509 |
83:0.822 |
||||
| chr14:71786965..71787090 | 16.831 | PCNX|pecanex homolog (Drosophila) | -412576 (+) | SIPA1L1|signal-induced proliferation-associated 1 like 1 | 209014 (+) | 6:0.951 |
|||||
| chr20:30945823..30945954 | 16.770 | KIF3B|kinesin family member 3B | -80402 (+) | ASXL1|additional sex combs like 1 (Drosophila) | 282 (+) | -13:0.681, -16:0.207 |
103:0.425 |
||||
| chr19:49999387..49999534 | 16.755 | RPL13A|ribosomal protein L13a | -8577 (+) | RPS11|ribosomal protein S11 | 187 (+) | 38:0.861, -41:0.574, -44:0.369 |
|||||
| chr9:98637809..98637943 | 16.707 | LINC00476|long intergenic non-protein coding RNA 476 | -23 (-) | C9orf102|chromosome 9 open reading frame 102 | 65 (+) | 11:0.805, 46:0.988, -90:0.557 |
|||||
| chr5:138629289..138629420 | 16.642 | MATR3|matrin 3 | -15 (+) | MATR3|matrin 3 | 105 (+) | 3:0.832 |
|||||
| chr15:22833356..22833486 | 16.631 | GOLGA6L1|golgin A6 family-like 1 | -97176 (+) | TUBGCP5|tubulin, gamma complex associated protein 5 | 49 (+) | 4:0.926, -47:0.964, 55:0.842 |
|||||
| chr19:8386252..8386365 | 16.622 | NDUFA7|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa | -22 (-) | RPS28|ribosomal protein S28 | 86 (+) | 13:0.917 |
|||||
| chr4:186064313..186064433 | 16.572 | HELT|helt bHLH transcription factor | -124335 (+) | SLC25A4|solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 | 97 (+) | 3:0.909 |
-37:0.624 |
||||
| chr6:35436079..35436194 | 16.505 | FANCE|Fanconi anemia, complementation group E | -15981 (+) | RPL10A|ribosomal protein L10a | 47 (+) | 0:0.441, -2:0.495 |
|||||
| chr17:53046022..53046135 | 16.488 | COX11|COX11 cytochrome c oxidase assembly homolog (yeast) | -61 (-) | STXBP4|syntaxin binding protein 4 | 49 (+) | 2:0.989, -19:0.894 |
|||||
| chr12:118454443..118454557 | 16.484 | KSR2|kinase suppressor of ras 2 | -48473 (-) | RFC5|replication factor C (activator 1) 5, 36.5kDa | 68 (+) | 8:0.983 |
|||||
| chr16:30709396..30709515 | 16.482 | FBRS|fibrosin | -33339 (+) | 481 (+) | 9:0.803 |
-75:0.957 |
|||||
| chr17:3627117..3627232 | 16.449 | ITGAE|integrin, alpha E (antigen CD103, human mucosal lymphocyte antigen 1; alpha polypeptide) | -72 (-) | GSG2|germ cell associated 2 (haspin) | 42 (+) | 20:0.907 |
|||||
| chr2:160568899..160569005 | 16.436 | BAZ2B|bromodomain adjacent to zinc finger domain, 2B | -46 (-) | MARCH7|membrane-associated ring finger (C3HC4) 7 | 43 (+) | -3:0.936 |
|||||
| chr16:30411985..30412099 | 16.330 | ZNF48|zinc finger protein 48 | -5301 (+) | ZNF771|zinc finger protein 771 | 6613 (+) | 0:0.316, 2:0.673 |
|||||
| chrX:30907498..30907629 | 16.325 | TAB3|TGF-beta activated kinase 1/MAP3K7 binding protein 3 | -104 (-) | FTHL17|ferritin, heavy polypeptide-like 17 | 182600 (-) | 7:0.891 |
|||||
| chr5:175788771..175788882 | 16.291 | KIAA1191|KIAA1191 | -63 (-) | ARL10|ADP-ribosylation factor-like 10 | 3663 (+) | 0:0.531, 50:0.388 |
36:0.935 |
||||
| chr5:139493593..139493688 | 16.286 | NRG2|neuregulin 2 | -70795 (-) | PURA|purine-rich element binding protein A | 87 (+) | -10:0.877 |
|||||
| chr17:4167249..4167354 | 16.277 | ANKFY1|ankyrin repeat and FYVE domain containing 1 | -150 (-) | UBE2G1|ubiquitin-conjugating enzyme E2G 1 | 102410 (-) | 15:0.878, -37:0.887 |
|||||
| chr3:49131487..49131599 | 16.273 | QRICH1|glutamine-rich 1 | -34 (-) | QARS|glutaminyl-tRNA synthetase | 10637 (-) | 5:0.734, 8:0.230, -15:0.884 |
-39:0.765 |
||||
| chr6:151711656..151711789 | 16.269 | AKAP12|A kinase (PRKA) anchor protein 12 | -64945 (+) | ZBTB2|zinc finger and BTB domain containing 2 | 988 (-) | 0:0.308, 17:0.466 |
|||||
| chr15:66648982..66649101 | 16.221 | TIPIN|TIMELESS interacting protein | -18 (-) | MAP2K1|mitogen-activated protein kinase kinase 1 | 30121 (+) | -2:0.939 |
-87:0.218 |
||||
| chr19:39936140..39936248 | 16.210 | RPS16|ribosomal protein S16 | -9618 (-) | SUPT5H|suppressor of Ty 5 homolog (S. cerevisiae) | 4 (+) | 6:0.826 |
|||||
| chr10:51565102..51565217 | 16.200 | NCOA4|nuclear receptor coactivator 4 | -20 (+) | NCOA4|nuclear receptor coactivator 4 | 44 (+) | -9:0.897, 41:0.492, 44:0.379 |
|||||
| chr1:156252624..156252748 | 16.193 | SMG5|smg-5 homolog, nonsense mediated mRNA decay factor (C. elegans) | -66 (-) | TMEM79|transmembrane protein 79 | 45 (+) | 7:0.808, -40:0.437, -43:0.443 |
|||||
| chr16:83841523..83841633 | 16.178 | CDH13|cadherin 13, H-cadherin (heart) | -1180909 (+) | HSBP1|heat shock factor binding protein 1 | 22 (+) | -5:0.299 |
-28:0.962 |
||||
| chr19:51014686..51014813 | 16.147 | JOSD2|Josephin domain containing 2 | -339 (-) | ASPDH|aspartate dehydrogenase domain containing | 2406 (-) | -7:0.929 |
|||||
| chr19:1095453..1095585 | 16.072 | POLR2E|polymerase (RNA) II (DNA directed) polypeptide E, 25kDa | -127 (-) | GPX4|glutathione peroxidase 4 (phospholipid hydroperoxidase) | 8451 (+) | 58:0.983 |
|||||
| chr17:74553737..74553862 | 16.055 | PRCD|progressive rod-cone degeneration | -17653 (+) | LOC100507246|uncharacterized LOC100507246 | 80 (+) | -2:0.304, -5:0.578 |
-30:0.973 |
||||
| chr15:85197502..85197616 | 16.047 | WDR73|WD repeat domain 73 | -33 (-) | NMB|neuromedin B | 3887 (-) | 8:0.629, 11:0.361 |
|||||
| chr16:87985023..87985159 | 16.040 | CA5A|carbonic anhydrase VA, mitochondrial | -14944 (-) | BANP|BTG3 associated nuclear protein | 77 (+) | -30:0.671 |
|||||
| chr6:108279349..108279467 | 16.020 | SEC63|SEC63 homolog (S. cerevisiae) | -16 (-) | SEC63|SEC63 homolog (S. cerevisiae) | 38 (-) | -2:0.977 |
|||||
| chr7:139044433..139044539 | 16.008 | C7orf55|chromosome 7 open reading frame 55 LUC7L2|LUC7-like 2 (S. cerevisiae) | -18392 (+) | LUC7L2|LUC7-like 2 (S. cerevisiae) | 148 (+) | -1:0.687, 1:0.276 |
|||||
| chr15:75315841..75315972 | 16.005 | SCAMP5|secretory carrier membrane protein 5 | -27999 (+) | PPCDC|phosphopantothenoylcysteine decarboxylase | 22 (+) | 21:0.633, 99:0.219 |
-65:0.991, -94:0.995 |
||||
| chr17:27717263..27717373 | 15.982 | NUFIP2|nuclear fragile X mental retardation protein interacting protein 2 | -96153 (-) | TAOK1|TAO kinase 1 | 627 (+) | -4:0.881 |
|||||
| chr17:4843497..4843630 | 15.962 | SLC25A11|solute carrier family 25 (mitochondrial carrier; oxoglutarate carrier), member 11 | -231 (-) | RNF167|ring finger protein 167 | 49 (+) | -2:0.477, -5:0.412, -44:0.223, -47:0.774, 55:0.896 |
36:0.206 |
||||
| chr16:57481271..57481378 | 15.938 | CCL17|chemokine (C-C motif) ligand 17 | -42629 (+) | CIAPIN1|cytokine induced apoptosis inhibitor 1 | 30 (-) | -7:0.439, -10:0.483 |
|||||
| chr10:69609182..69609284 | 15.903 | DNAJC12|DnaJ (Hsp40) homolog, subfamily C, member 12 | -11381 (-) | SIRT1|sirtuin 1 | 35174 (+) | 3:0.674 |
-20:0.993 |
28:0.332 |
|||
| chr2:677398..677540 | 15.810 | TMEM18|transmembrane protein 18 | -58 (-) | SNTG2|syntrophin, gamma 2 | 269110 (+) | 4:0.911, 36:0.430, -38:0.706, 39:0.524 |
|||||
| chr4:15683195..15683323 | 15.798 | FBXL5|F-box and leucine-rich repeat protein 5 | -26244 (-) | FAM200B|family with sequence similarity 200, member B | 67 (+) | 6:0.984, -11:0.986 |
|||||
| chr16:67867798..67867908 | 15.765 | CENPT|centromere protein T | -24 (-) | THAP11|THAP domain containing 11 | 8362 (+) | -8:0.524, -11:0.343 |
17:0.982 |
||||
| chr19:19431538..19431651 | 15.747 | SUGP1|SURP and G patch domain containing 1 | -243 (-) | MAU2|MAU2 chromatid cohesion factor homolog (C. elegans) | 40 (+) | -40:0.962 |
|||||
| chr14:103800271..103800369 | 15.712 | TNFAIP2|tumor necrosis factor, alpha-induced protein 2 | -207552 (+) | EIF5|eukaryotic translation initiation factor 5 | 190 (+) | -8:0.495, -11:0.495 |
|||||
| chr17:78428523..78428636 | 15.627 | ENDOV|endonuclease V | -39329 (+) | NPTX1|neuronal pentraxin I | 21748 (-) | -3:0.983 |
|||||
| chr9:139780742..139780858 | 15.622 | EDF1|endothelial differentiation-related factor 1 | -20062 (-) | TRAF2|TNF receptor-associated factor 2 | 153 (+) | -40:0.913 |
|||||
| chr2:96931798..96931935 | 15.614 | TMEM127|transmembrane protein 127 | -111 (-) | CIAO1|cytosolic iron-sulfur protein assembly 1 | 42 (+) | -4:0.202 |
-26:0.953, 31:0.998 |
||||
| chr2:225450204..225450310 | 15.610 | CUL3|cullin 3 | -137 (-) | DOCK10|dedicator of cytokinesis 10 | 361497 (-) | -11:0.991 |
|||||
| chr19:7894844..7894993 | 15.598 | CLEC4M|C-type lectin domain family 4, member M | -66825 (+) | EVI5L|ecotropic viral integration site 5-like | 258 (+) | ||||||
| chr1:45987545..45987660 | 15.593 | PRDX1|peroxiredoxin 1 | -24 (-) | PRDX1|peroxiredoxin 1 | 959 (-) | 0:0.537, 2:0.457 |
|||||
| chr1:179923791..179923914 | 15.577 | TOR1AIP1|torsin A interacting protein 1 | -71866 (+) | CEP350|centrosomal protein 350kDa | 53 (+) | 8:0.885 |
|||||
| chr16:87526624..87526736 | 15.569 | ZCCHC14|zinc finger, CCHC domain containing 14 | -1290 (-) | JPH3|junctophilin 3 | 109821 (+) | -26:0.887 |
12:0.788 |
||||
| chr10:27149948..27150084 | 15.513 | ABI1|abl-interactor 1 | -11 (-) | ANKRD26|ankyrin repeat domain 26 | 239368 (-) | -13:0.906 |
54:0.996 |
||||
| chr3:139108463..139108570 | 15.512 | -19 (-) | COPB2|coatomer protein complex, subunit beta 2 (beta prime) | 3 (-) | 0:0.852 |
||||||
| chr3:194304608..194304730 | 15.494 | FLJ34208|uncharacterized LOC401106 | -96867 (+) | 64 (+) | |||||||
| chr7:100472546..100472701 | 15.494 | TRIP6|thyroid hormone receptor interactor 6 | -7589 (+) | SRRT|serrate RNA effector molecule homolog (Arabidopsis) | 73 (+) | 5:0.556 |
83:0.210 |
||||
| chr12:125473627..125473744 | 15.468 | DHX37|DEAH (Asp-Glu-Ala-His) box polypeptide 37 | -41 (-) | BRI3BP|BRI3 binding protein | 4505 (+) | 0:0.412, 2:0.539 |
|||||
| chr3:169684485..169684605 | 15.468 | SAMD7|sterile alpha motif domain containing 7 | -55125 (+) | SEC62|SEC62 homolog (S. cerevisiae) | 43 (+) | 7:0.972 |
|||||
| chr10:70091694..70091810 | 15.425 | PBLD|phenazine biosynthesis-like protein domain containing | -25138 (-) | HNRNPH3|heterogeneous nuclear ribonucleoprotein H3 (2H9) | 70 (+) | -7:0.927 |
|||||
| chr4:17812478..17812590 | 15.394 | DCAF16|DDB1 and CUL4 associated factor 16 | -261 (-) | NCAPG|non-SMC condensin I complex, subunit G | 77 (+) | 7:0.984, 25:0.971 |
|||||
| chr2:25194911..25195032 | 15.384 | DNAJC27|DnaJ (Hsp40) homolog, subfamily C, member 27 | -104 (-) | EFR3B|EFR3 homolog B (S. cerevisiae) | 70006 (+) | 4:0.707 |
26:0.238 |
||||
| chr6:100016666..100016776 | 15.374 | CCNC|cyclin C | -118 (-) | PRDM13|PR domain containing 13 | 37928 (+) | 10:0.583, 13:0.409 |
|||||
| chr19:42364221..42364341 | 15.370 | RPS19|ribosomal protein S19 | -5 (+) | RPS19|ribosomal protein S19 | 49 (+) | -1:0.865, -17:0.873 |
|||||
| chr21:30446031..30446147 | 15.366 | CCT8|chaperonin containing TCP1, subunit 8 (theta) | -100 (-) | C21orf7|chromosome 21 open reading frame 7 | 6783 (+) | -19:0.943 |
|||||
| chr6:47445419..47445547 | 15.360 | TNFRSF21|tumor necrosis factor receptor superfamily, member 21 | -167788 (-) | CD2AP|CD2-associated protein | 32 (+) | -1:0.956 |
|||||
| chr9:88555876..88556000 | 15.341 | AGTPBP1|ATP/GTP binding protein 1 | -199067 (-) | NAA35|N(alpha)-acetyltransferase 35, NatC auxiliary subunit | 136 (+) | 2:0.967, -12:0.489 |
|||||
| chr1:6296047..6296138 | 15.339 | ICMT|isoprenylcysteine carboxyl methyltransferase | -82 (-) | ICMT|isoprenylcysteine carboxyl methyltransferase | 16 (-) | 17:0.498, 20:0.498 |
|||||
| chr7:129845277..129845385 | 15.333 | TMEM209|transmembrane protein 209 | -55 (-) | C7orf45|chromosome 7 open reading frame 45 | 2377 (+) | 7:0.309, 10:0.575 |
|||||
| chr6:97731102..97731230 | 15.319 | MMS22L|MMS22-like, DNA repair protein | -76 (-) | POU3F2|POU class 3 homeobox 2 | 1551414 (+) | 9:0.668, 12:0.328, -28:0.911 |
|||||
| chr17:15602827..15602963 | 15.295 | TRIM16|tripartite motif containing 16 | -16703 (-) | ZNF286A|zinc finger protein 286A | 68 (+) | -21:0.943 |
84:0.997 |
||||
| chr6:52284941..52285069 | 15.285 | PAQR8|progestin and adipoQ receptor family member VIII | -58048 (+) | EFHC1|EF-hand domain (C-terminal) containing 1 | 73 (+) | 2:0.433 |
|||||
| chr6:36853541..36853671 | 15.259 | PPIL1|peptidylprolyl isomerase (cyclophilin)-like 1 | -10806 (-) | C6orf89|chromosome 6 open reading frame 89 | 104 (+) | -14:0.974 |
|||||
| chr13:41706962..41707100 | 15.245 | KBTBD6|kelch repeat and BTB (POZ) domain containing 6 | -236 (-) | KBTBD7|kelch repeat and BTB (POZ) domain containing 7 | 61426 (-) | 43:0.214 |
22:0.900, -71:0.981 |
||||
| chr2:220363521..220363641 | 15.245 | SPEG|SPEG complex locus | -38048 (+) | GMPPA|GDP-mannose pyrophosphorylase A | 200 (+) | -25:0.993 |
|||||
| chr15:78730463..78730573 | 15.233 | CRABP1|cellular retinoic acid binding protein 1 | -97815 (+) | IREB2|iron-responsive element binding protein 2 | 3 (+) | -8:0.845, -28:0.974, 29:0.961 |
|||||
| chr15:64126141..64126253 | 15.230 | HERC1|hect (homologous to the E6-AP (UBE3A) carboxyl terminus) domain and RCC1 (CHC1)-like domain (RLD) 1 | -57 (-) | DAPK2|death-associated protein kinase 2 | 212413 (-) | -5:0.793 |
|||||
| chr7:2272542..2272639 | 15.230 | MAD1L1|MAD1 mitotic arrest deficient-like 1 (yeast) | -10 (-) | FTSJ2|FtsJ homolog 2 (E. coli) | 9217 (-) | 10:0.829, -12:0.716 |
|||||
| chr21:47604481..47604624 | 15.226 | C21orf56|chromosome 21 open reading frame 56 | -177 (-) | LSS|lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) | 43953 (-) | ||||||
| chr17:27279378..27279496 | 15.219 | PHF12|PHD finger protein 12 | -941 (-) | SEZ6|seizure related 6 homolog (mouse) | 53588 (-) | 25:0.956, -70:0.942 |
|||||
| chr17:37607517..37607655 | 15.219 | MED1|mediator complex subunit 1 | -165 (-) | CDK12|cyclin-dependent kinase 12 | 10166 (+) | 25:0.840, -60:0.218 |
|||||
| chr1:27226906..27227023 | 15.209 | GPATCH3|G patch domain containing 3 | -13 (-) | NR0B2|nuclear receptor subfamily 0, group B, member 2 | 13534 (-) | 5:0.905 |
|||||
| chr5:56469802..56469925 | 15.187 | GPBP1|GC-rich promoter binding protein 1 | -149 (+) | GPBP1|GC-rich promoter binding protein 1 | 34 (+) | 6:0.806 |
|||||
| chr5:127418753..127418861 | 15.184 | FLJ33630|uncharacterized LOC644873 | -44 (-) | FLJ33630|uncharacterized LOC644873 | 552 (-) | 2:0.981 |
|||||
| chr17:55162323..55162449 | 15.175 | SCPEP1|serine carboxypeptidase 1 | -106882 (+) | AKAP1|A kinase (PRKA) anchor protein 1 | 228 (+) | 0:0.803 |
|||||
| chr7:35840539..35840658 | 15.168 | HERPUD2|HERPUD family member 2 | -105820 (-) | SEPT7|septin 7 | 112 (+) | -4:0.936 |
|||||
| chr7:100303570..100303701 | 15.150 | GIGYF1|GRB10 interacting GYF protein 1 | -16766 (-) | POP7|processing of precursor 7, ribonuclease P/MRP subunit (S. cerevisiae) | 85 (+) | ||||||
| chr10:101989321..101989443 | 15.148 | CHUK|conserved helix-loop-helix ubiquitous kinase | -24 (-) | CWF19L1|CWF19-like 1, cell cycle control (S. pombe) | 38004 (-) | -5:0.476, -8:0.507 |
|||||
| chr19:14228645..14228766 | 15.138 | PRKACA|protein kinase, cAMP-dependent, catalytic, alpha | -136 (-) | ASF1B|ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | 18627 (-) | 22:0.284 |
54:0.744, -63:0.819 |
||||
| chr15:99791465..99791579 | 15.136 | TTC23|tetratricopeptide repeat domain 23 | -106 (-) | LRRC28|leucine rich repeat containing 28 | 120 (+) | 24:0.259, 27:0.488 |
|||||
| chr12:57940962..57941099 | 15.125 | DCTN2|dynactin 2 (p50) | -102 (-) | KIF5A|kinesin family member 5A | 2760 (+) | -12:0.705, -48:0.531, -51:0.403 |
92:0.405 |
||||
| chr16:4666184..4666339 | 15.095 | FAM100A|family with sequence similarity 100, member A | -1233 (-) | MGRN1|mahogunin, ring finger 1 | 8562 (+) | 99:0.487 |
24:0.672 |
70:0.242 |
|||
| chr6:170102122..170102237 | 15.064 | WDR27|WD repeat domain 27 | -22 (-) | C6orf120|chromosome 6 open reading frame 120 | 40 (+) | -8:0.966 |
48:0.741 |
||||
| chr13:22178297..22178419 | 15.061 | EFHA1|EF-hand domain family, member A1 | -49 (-) | EFHA1|EF-hand domain family, member A1 | 2 (-) | -1:0.596, -4:0.357 |
|||||
| chr13:20532716..20532838 | 15.057 | ZMYM5|zinc finger, MYM-type 5 | -94969 (-) | ZMYM2|zinc finger, MYM-type 2 | 40 (+) | -13:0.821, 30:0.950 |
|||||
| chr16:188614..188737 | 15.047 | NPRL3|nitrogen permease regulator-like 3 (S. cerevisiae) | -221 (-) | NPRL3|nitrogen permease regulator-like 3 (S. cerevisiae) | 4 (-) | -10:0.842 |
|||||
| chr10:127408079..127408255 | 15.021 | C10orf137|chromosome 10 open reading frame 137 | -54 (+) | C10orf137|chromosome 10 open reading frame 137 | 145 (+) | 56:0.688, 59:0.211, -60:0.962 |
-10:0.875 |
||||
| chr11:8704194..8704326 | 15.011 | RPL27A|ribosomal protein L27a | -266 (+) | RPL27A|ribosomal protein L27a | 80 (+) | -26:0.909, 56:0.414, 59:0.564 |
|||||
| chr2:170683866..170683978 | 15.010 | METTL5|methyltransferase like 5 | -2537 (-) | UBR3|ubiquitin protein ligase E3 component n-recognin 3 (putative) | 36 (+) | 0:0.628, -2:0.350 |
|||||
| chr3:49044706..49044835 | 14.989 | WDR6|WD repeat domain 6 | -154 (+) | WDR6|WD repeat domain 6 | 121 (+) | 41:0.938 |
|||||
| chr1_gl000191_random:50148..50296 | 14.988 | None | None (None) | None | None (None) | -27:0.987, 40:0.424 |
|||||
| chr1:43824471..43824598 | 14.968 | MPL|myeloproliferative leukemia virus oncogene | -21038 (+) | CDC20|cell division cycle 20 homolog (S. cerevisiae) | 208 (+) | -29:0.336 |
20:0.385, 65:0.572 |
||||
| chr15:78730341..78730450 | 14.941 | CRABP1|cellular retinoic acid binding protein 1 | -97692 (+) | IREB2|iron-responsive element binding protein 2 | 126 (+) | 8:0.555 |
|||||
| chr17:7760861..7760999 | 14.938 | LSMD1|LSM domain containing 1 | -110 (-) | CYB5D1|cytochrome b5 domain containing 1 | 133 (+) | 0:0.769, -49:0.597, 69:0.859, -76:0.375, -79:0.597 |
|||||
| chr12:510623..510750 | 14.935 | KDM5A|lysine (K)-specific demethylase 5A | -12084 (-) | CCDC77|coiled-coil domain containing 77 | 95 (+) | 28:0.411 |
-53:0.992 |
||||
| chr19:13906031..13906169 | 14.927 | C19orf53|chromosome 19 open reading frame 53 | -20816 (+) | ZSWIM4|zinc finger, SWIM-type containing 4 | 145 (+) | 59:0.591, -69:0.890, 96:0.364 |
|||||
| chr11:64646239..64646338 | 14.918 | EHD1|EH-domain containing 1 | -134 (-) | EHD1|EH-domain containing 1 | 873 (-) | -17:0.814 |
-35:0.840 |
||||
| chr6:146285542..146285670 | 14.914 | SHPRH|SNF2 histone linker PHD RING helicase | -44 (-) | GRM1|glutamate receptor, metabotropic 1 | 63243 (+) | 11:0.930 |
|||||
| chr18:47018848..47018950 | 14.913 | RPL17-C18ORF32|RPL17-C18orf32 readthrough RPL17|ribosomal protein L17 | -11 (-) | LIPG|lipase, endothelial | 68163 (+) | -1:0.895 |
|||||
| chr14:100705085..100705217 | 14.903 | YY1|YY1 transcription factor | -9 (+) | YY1|YY1 transcription factor | 161 (+) | 8:0.747, 11:0.225 |
|||||
| chr8:42698435..42698572 | 14.875 | THAP1|THAP domain containing, apoptosis associated protein 1 | -42 (-) | RNF170|ring finger protein 170 | 53240 (-) | -19:0.616, -22:0.273 |
|||||
| chr19:1652917..1653070 | 14.848 | TCF3|transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | -654 (-) | ONECUT3|one cut homeobox 3 | 100759 (+) | 78:0.359 |
|||||
| chr12:54020127..54020263 | 14.847 | ATF7|activating transcription factor 7 | -42 (-) | ATP5G2|ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C2 (subunit 9) | 49681 (-) | -7:0.963 |
|||||
| chr9:6681650..6681759 | 14.846 | GLDC|glycine dehydrogenase (decarboxylating) | -36077 (-) | KDM4C|lysine (K)-specific demethylase 4C | 39158 (+) | -6:0.443, -9:0.549 |
|||||
| chr1:24306821..24306967 | 14.834 | SRSF10|serine/arginine-rich splicing factor 10 | -2 (-) | MYOM3|myomesin family, member 3 | 128075 (-) | -27:0.987, 40:0.424 |
|||||
| chr1:247095281..247095403 | 14.827 | AHCTF1P1|AT hook containing transcription factor 1 pseudogene 1 AHCTF1|AT hook containing transcription factor 1 | -650 (-) | ZNF695|zinc finger protein 695 | 75970 (-) | 7:0.913 |
|||||
| chrX:153607340..153607494 | 14.824 | FLNA|filamin A, alpha | -4416 (-) | EMD|emerin | 196 (+) | 64:0.210 |
17:0.361, -83:0.361 |
||||
| chr8:37594032..37594163 | 14.809 | ZNF703|zinc finger protein 703 | -40780 (+) | ERLIN2|ER lipid raft associated 2 | 9 (+) | -15:0.759 |
|||||
| chrX:70474024..70474140 | 14.794 | ZMYM3|zinc finger, MYM-type 3 | -74 (-) | ZMYM3|zinc finger, MYM-type 3 | 404 (-) | 3:0.638 |
-19:0.657 |
||||
| chr5:176449599..176449731 | 14.790 | UIMC1|ubiquitin interaction motif containing 1 | -16077 (-) | ZNF346|zinc finger protein 346 | 54 (+) | -16:0.305, -19:0.688, 22:0.370, 25:0.621 |
|||||
| chr2:55844796..55844927 | 14.769 | SMEK2|SMEK homolog 2, suppressor of mek1 (Dictyostelium) | -67 (-) | SMEK2|SMEK homolog 2, suppressor of mek1 (Dictyostelium) | 50 (-) | 11:0.752 |
|||||
| chr6:44355199..44355316 | 14.763 | SPATS1|spermatogenesis associated, serine-rich 1 | -44817 (+) | CDC5L|CDC5 cell division cycle 5-like (S. pombe) | 15 (+) | -5:0.806 |
|||||
| chr19:1438297..1438417 | 14.761 | DAZAP1|DAZ associated protein 1 | -30590 (+) | RPS15|ribosomal protein S15 | 8 (+) | 17:0.893 |
|||||
| chr14:104181800..104181976 | 14.758 | XRCC3|X-ray repair complementing defective repair in Chinese hamster cells 3 | -105 (-) | ZFYVE21|zinc finger, FYVE domain containing 21 | 228 (+) | 7:0.237, -34:0.568, 68:0.605 |
|||||
| chr19:5903742..5903873 | 14.745 | NDUFA11|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa | -47 (-) | NDUFA11|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 11, 14.7kDa | 200 (-) | -12:0.527, -15:0.449 |
|||||
| chr15:69452847..69452961 | 14.721 | GLCE|glucuronic acid epimerase | -6 (+) | PAQR5|progestin and adipoQ receptor family member V | 138384 (+) | 0:0.488 |
|||||
| chr20:30467899..30467997 | 14.707 | TTLL9|tubulin tyrosine ligase-like family, member 9 | -335 (+) | PDRG1|p53 and DNA-damage regulated 1 | 71812 (-) | -2:0.758 |
-45:0.992 |
||||
| chr2:85555073..85555188 | 14.693 | TGOLN2|trans-golgi network protein 2 | 0 (-) | TGOLN2|trans-golgi network protein 2 | 258 (-) | -6:0.607, -9:0.386 |
|||||
| chr15:76603768..76603885 | 14.685 | ETFA|electron-transfer-flavoprotein, alpha polypeptide | -12 (-) | ISL2|ISL LIM homeobox 2 | 25336 (+) | -12:0.861 |
55:0.441, 74:0.331 |
||||
| chr17:65713930..65714060 | 14.671 | PITPNC1|phosphatidylinositol transfer protein, cytoplasmic 1 | -340072 (+) | NOL11|nucleolar protein 11 | 55 (+) | 9:0.648, 12:0.327 |
|||||
| chr1:53308426..53308552 | 14.668 | ZYG11A|zyg-11 homolog A (C. elegans) | -307 (+) | ECHDC2|enoyl CoA hydratase domain containing 2 | 78738 (-) | 7:0.932 |
|||||
| chr1:28052113..28052223 | 14.664 | IFI6|interferon, alpha-inducible protein 6 | -53429 (-) | FAM76A|family with sequence similarity 76, member A | 360 (+) | -12:0.982 |
|||||
| chr19:14117163..14117334 | 14.642 | RFX1|regulatory factor X, 1 (influences HLA class II expression) | -119 (-) | RLN3|relaxin 3 | 21739 (+) | -65:0.230 |
|||||
| chr9:136214985..136215092 | 14.640 | MED22|mediator complex subunit 22 | -76 (-) | RPL7A|ribosomal protein L7a | 37 (+) | -5:0.345, -8:0.389 |
|||||
| chr4:178363602..178363727 | 14.639 | AGA|aspartylglucosaminidase | -21 (-) | ODZ3|odz, odd Oz/ten-m homolog 3 (Drosophila) | 4701461 (+) | 1:0.984 |
|||||
| chr17:78121038..78121168 | 14.633 | EIF4A3|eukaryotic translation initiation factor 4A3 | -233 (-) | CARD14|caspase recruitment domain family, member 14 | 31185 (+) | -1:0.297, 16:0.489 |
|||||
| chr2:176032860..176032969 | 14.631 | ATF2|activating transcription factor 2 | -19 (-) | ATP5G3|ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C3 (subunit 9) | 13390 (-) | 3:0.854 |
|||||
| chr3:184016731..184016834 | 14.620 | ECE2|endothelin converting enzyme 2 | -22914 (+) | PSMD2|proteasome (prosome, macropain) 26S subunit, non-ATPase, 2 | 228 (+) | 54:0.996 |
|||||
| chr17:8286501..8286636 | 14.587 | RPL26|ribosomal protein L26 | -64 (-) | RNF222|ring finger protein 222 | 14575 (-) | -11:0.360, -14:0.638 |
|||||
| chr1:207226278..207226408 | 14.584 | YOD1|YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae) | -1984 (-) | PFKFB2|6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 | 294 (+) | -8:0.520, -11:0.474 |
|||||
| chr12:123849752..123849877 | 14.579 | SBNO1|strawberry notch homolog 1 (Drosophila) | -49 (-) | SETD8|SET domain containing (lysine methyltransferase) 8 | 18841 (+) | -18:0.346 |
-47:0.246 |
||||
| chr11:67981324..67981434 | 14.566 | SUV420H1|suppressor of variegation 4-20 homolog 1 (Drosophila) | -400 (-) | C11orf24|chromosome 11 open reading frame 24 | 58007 (-) | ||||||
| chr3:14220258..14220381 | 14.563 | LSM3|LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | -45 (+) | LSM3|LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 15 (+) | 7:0.799 |
|||||
| chr12:29534112..29534244 | 14.562 | ERGIC2|ERGIC and golgi 2 | -63 (-) | OVCH1|ovochymase 1 | 116440 (-) | 8:0.487, 11:0.498, -52:0.978 |
|||||
| chr15:40075013..40075121 | 14.533 | FSIP1|fibrous sheath interacting protein 1 | -39 (-) | 39 (+) | -9:0.771 |
||||||
| chr17:79196799..79196931 | 14.495 | AZI1|5-azacytidine induced 1 | -99 (-) | C17orf56|chromosome 17 open reading frame 56 | 15958 (-) | 13:0.232, -27:0.399, 98:0.243 |
|||||
| chr19:2944970..2945135 | 14.447 | ZNF77|zinc finger protein 77 | -109 (-) | TLE6|transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) | 32514 (+) | -1:0.244, -32:0.759, -63:0.340, 71:0.759 |
|||||
| chr8:33342569..33342690 | 14.447 | FUT10|fucosyltransferase 10 (alpha (1,3) fucosyltransferase) | -11983 (-) | MAK16|MAK16 homolog (S. cerevisiae) | 75 (+) | ||||||
| chr3:137893723..137893837 | 14.444 | DBR1|debranching enzyme homolog 1 (S. cerevisiae) | -11 (-) | ARMC8|armadillo repeat containing 8 | 12563 (+) | 13:0.973 |
|||||
| chr1:1310651..1310815 | 14.432 | AURKAIP1|aurora kinase A interacting protein 1 | -160 (-) | AURKAIP1|aurora kinase A interacting protein 1 | 130 (-) | 60:0.973 |
|||||
| chr4:128886276..128886392 | 14.431 | MFSD8|major facilitator superfamily domain containing 8 | -19 (-) | C4orf29|chromosome 4 open reading frame 29 | 122 (+) | -4:0.782 |
|||||
| chr1:1051518..1051675 | 14.430 | C1orf159|chromosome 1 open reading frame 159 | -120 (-) | C1orf159|chromosome 1 open reading frame 159 | 139 (-) | -10:0.300 |
-25:0.996, 72:0.991, -87:0.996 |
||||
| chr22:43010907..43011019 | 14.425 | POLDIP3|polymerase (DNA-directed), delta interacting protein 3 | -12 (-) | 16482 (-) | 3:0.435, 6:0.508, -61:0.494 |
||||||
| chr6:144164346..144164466 | 14.425 | PHACTR2|phosphatase and actin regulator 2 | -165229 (+) | LTV1|LTV1 homolog (S. cerevisiae) | 83 (+) | -16:0.434 |
44:0.586 |
||||
| chr1:110577109..110577192 | 14.402 | AHCYL1|adenosylhomocysteinase-like 1 | -17023 (+) | FAM40A|family with sequence similarity 40, member A | 79 (+) | -33:0.272, -36:0.551, 49:0.843 |
|||||
| chr17:26368557..26368708 | 14.382 | C17orf108|chromosome 17 open reading frame 108 | -148241 (-) | NLK|nemo-like kinase | 1021 (+) | -67:0.514, 73:0.271 |
|||||
| chr5:132387577..132387701 | 14.381 | HSPA4|heat shock 70kDa protein 4 | -1 (+) | HSPA4|heat shock 70kDa protein 4 | 62 (+) | -1:0.893 |
|||||
| chr16:75657208..75657333 | 14.345 | ADAT1|adenosine deaminase, tRNA-specific 1 | -84 (-) | KARS|lysyl-tRNA synthetase | 24291 (-) | -14:0.800 |
|||||
| chr22:45809463..45809589 | 14.344 | SMC1B|structural maintenance of chromosomes 1B | -42 (-) | RIBC2|RIB43A domain with coiled-coils 2 | 43 (+) | -2:0.973 |
|||||
| chr16:4818733..4818874 | 14.335 | ZNF500|zinc finger protein 500 | -1619 (-) | SEPT12|septin 12 | 19615 (-) | -7:0.258 |
77:0.991 |
||||
| chr15:73925714..73925844 | 14.328 | NPTN|neuroplastin | -53 (-) | 50769 (+) | 5:0.792, -45:0.303, -48:0.683 |
76:0.521 |
|||||
| chr12:11324200..11324314 | 14.320 | PRH1-PRR4|PRH1-PRR4 readthrough PRH1|proline-rich protein HaeIII subfamily 1 TAS2R14|taste receptor, type 2, member 14 | -65 (-) | TAS2R42|taste receptor, type 2, member 42 | 15285 (-) | 5:0.443, 8:0.548 |
-13:0.578 |
||||
| chr9:86595513..86595659 | 14.318 | HNRNPK|heterogeneous nuclear ribonucleoprotein K | -47 (-) | RMI1|RMI1, RecQ mediated genome instability 1, homolog (S. cerevisiae) | 123 (+) | -1:0.937 |
|||||
| chr10:5932014..5932139 | 14.305 | ANKRD16|ankyrin repeat domain 16 | -220 (-) | FBXO18|F-box protein, helicase, 18 | 123 (+) | 28:0.848 |
|||||
| chr16:84150395..84150527 | 14.296 | MBTPS1|membrane-bound transcription factor peptidase, site 1 | -49 (-) | MBTPS1|membrane-bound transcription factor peptidase, site 1 | 55 (-) | -20:0.882 |
|||||
| chr3:169482875..169482988 | 14.290 | TERC|telomerase RNA component | -83 (-) | ARPM1|actin related protein M1 | 4711 (-) | -1:0.313, 1:0.228 |
-27:0.257 |
||||
| chr12:72057615..72057752 | 14.287 | ZFC3H1|zinc finger, C3H1-type containing | -85 (-) | ZFC3H1|zinc finger, C3H1-type containing | 29 (-) | -44:0.209, -47:0.334 |
67:0.703 |
||||
| chr9:140135495..140135664 | 14.285 | SLC34A3|solute carrier family 34 (sodium phosphate), member 3 | -10195 (+) | TUBB4B|tubulin, beta 4B class IVb | 129 (+) | -102:0.792 |
|||||
| chr16:2014815..2014968 | 14.280 | RPS2|ribosomal protein S2 | -63 (-) | RNF151|ring finger protein 151 | 1983 (+) | -20:0.627 |
43:0.857 |
||||
| chr2:55844942..55845057 | 14.277 | SMEK2|SMEK homolog 2, suppressor of mek1 (Dictyostelium) | -88 (-) | SMEK2|SMEK homolog 2, suppressor of mek1 (Dictyostelium) | 1019 (-) | 0:0.535 |
|||||
| chr4:103749104..103749280 | 14.266 | UBE2D3|ubiquitin-conjugating enzyme E2D 3 | -56 (-) | UBE2D3|ubiquitin-conjugating enzyme E2D 3 | 40824 (-) | 39:0.958, -65:0.623, -68:0.373 |
|||||
| chr4:26321188..26321300 | 14.250 | C4orf52|chromosome 4 open reading frame 52 | -405380 (+) | RBPJ|recombination signal binding protein for immunoglobulin kappa J region | 75 (+) | -4:0.972 |
|||||
| chr10:64892827..64892971 | 14.233 | EGR2|early growth response 2 | -313973 (-) | NRBF2|nuclear receptor binding factor 2 | 132 (+) | -8:0.590, -11:0.306 |
|||||
| chr7:100184129..100184257 | 14.232 | FBXO24|F-box protein 24 | -191 (+) | FBXO24|F-box protein 24 | 3030 (+) | -9:0.416, -50:0.229 |
|||||
| chr4:140374838..140374955 | 14.207 | -22 (-) | RAB33B|RAB33B, member RAS oncogene family | 64 (+) | -4:0.662, -7:0.274 |
||||||
| chr5:138940691..138940827 | 14.202 | TMEM173|transmembrane protein 173 | -78416 (-) | UBE2D2|ubiquitin-conjugating enzyme E2D 2 | 14 (+) | -23:0.499, -26:0.499 |
|||||
| chr12:67662898..67663026 | 14.201 | GRIP1|glutamate receptor interacting protein 1 | -590060 (-) | CAND1|cullin-associated and neddylation-dissociated 1 | 140 (+) | -27:0.301, -30:0.689 |
|||||
| chr1:111991879..111992008 | 14.199 | WDR77|WD repeat domain 77 | -74 (-) | ATP5F1|ATP synthase, H+ transporting, mitochondrial Fo complex, subunit B1 | 116 (+) | -19:0.924, 63:0.975 |
|||||
| chr12:27091272..27091439 | 14.198 | ASUN|asunder, spermatogenesis regulator homolog (Drosphila) | -125 (-) | FGFR1OP2|FGFR1 oncogene partner 2 | 20 (+) | 37:0.814, -97:0.206, -100:0.752 |
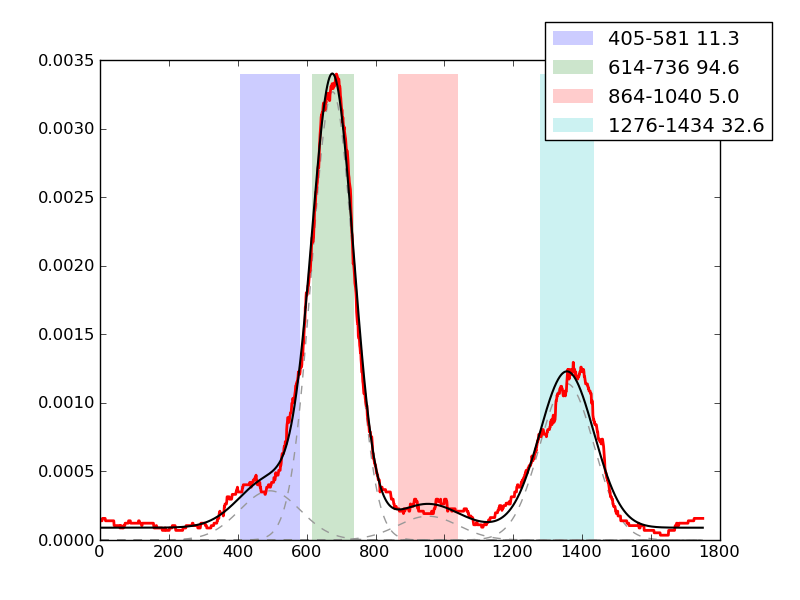
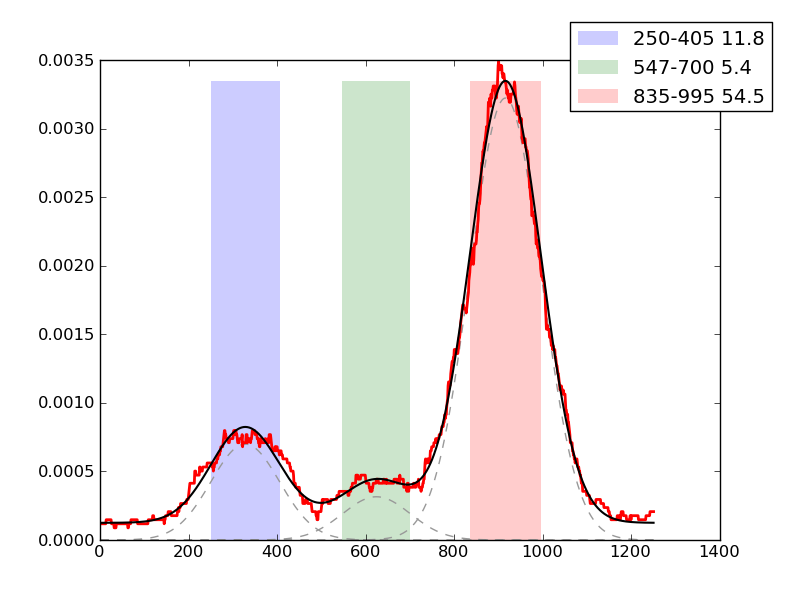
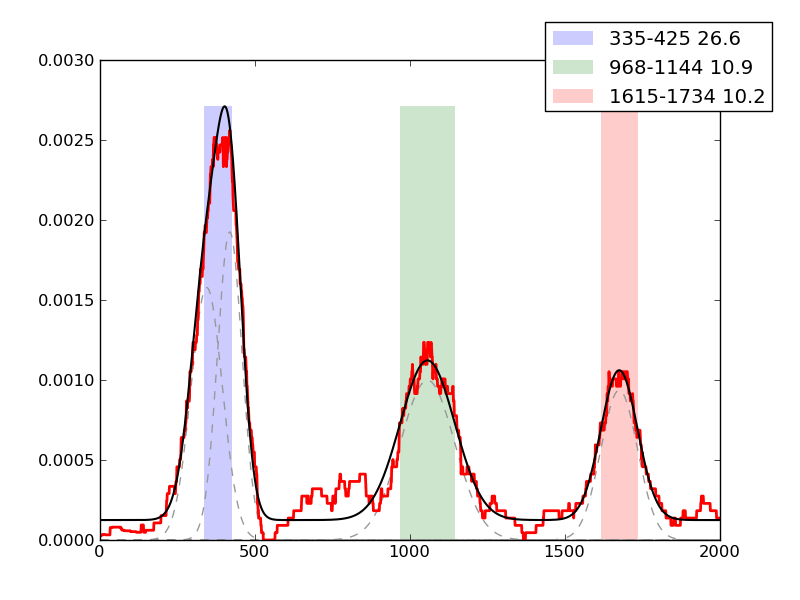
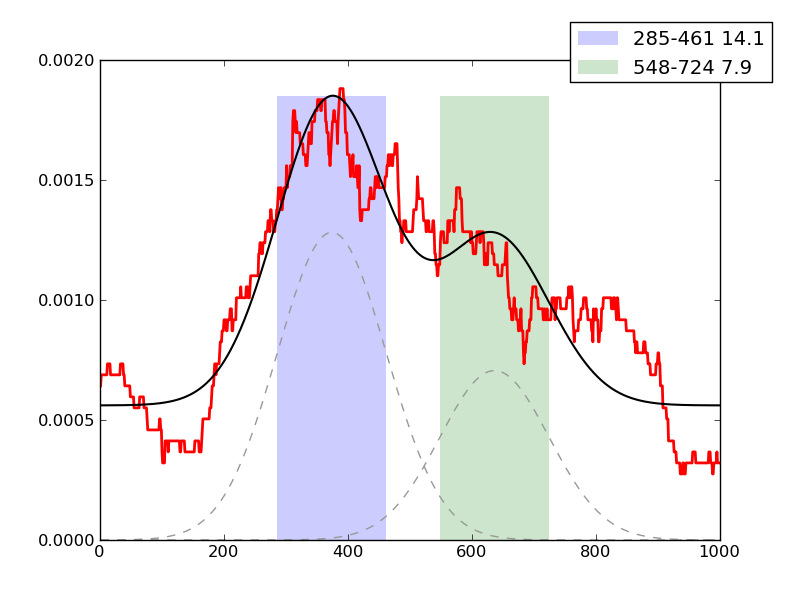
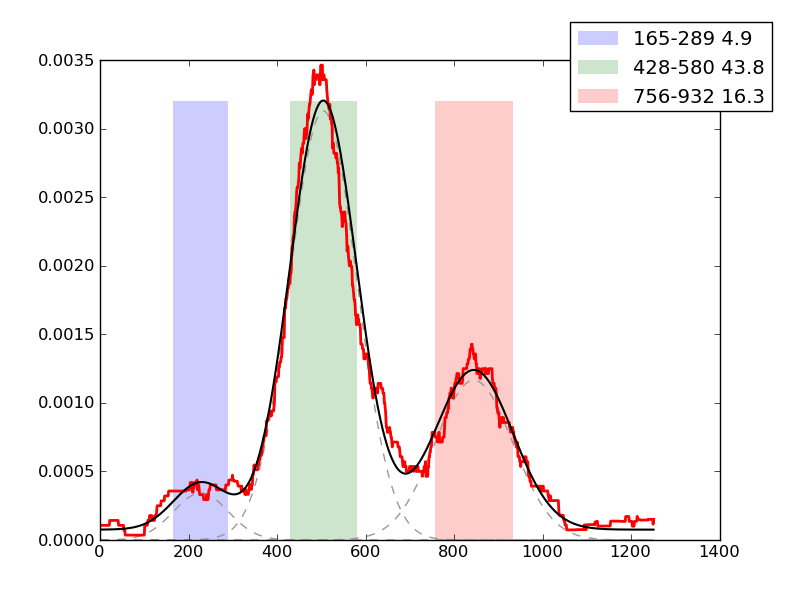
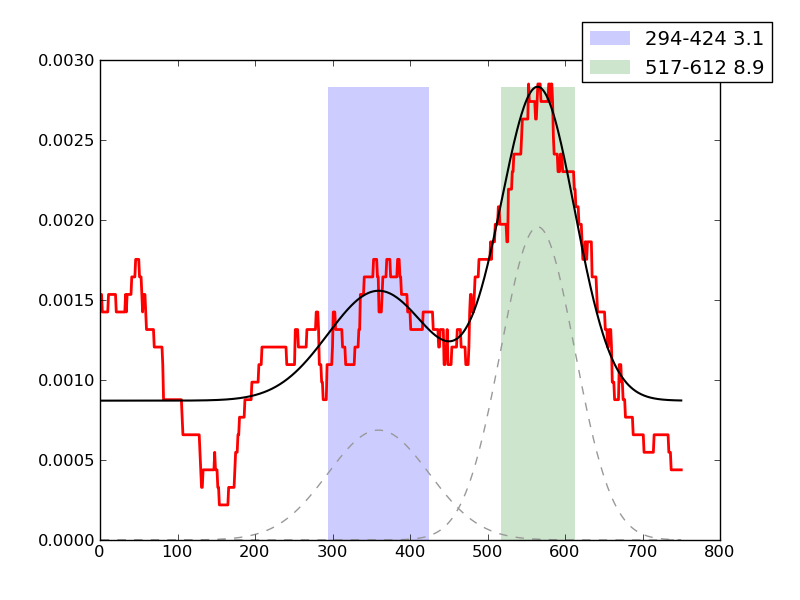
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| denovo_WM_21 | 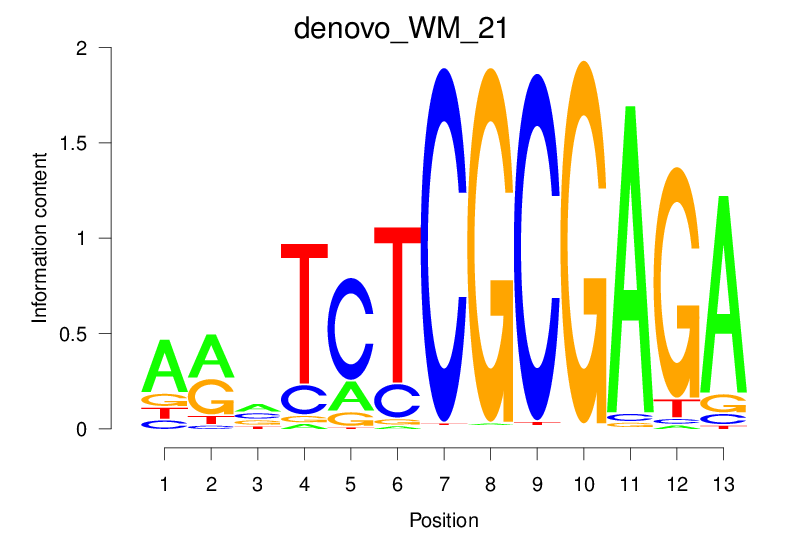 |
2.951 (541.109) | 2.951 (541.109) | 0.5226 | 0.4347 | 17.0817 | 1334/37435 |
| HCMC.NFYA_f1.wm | 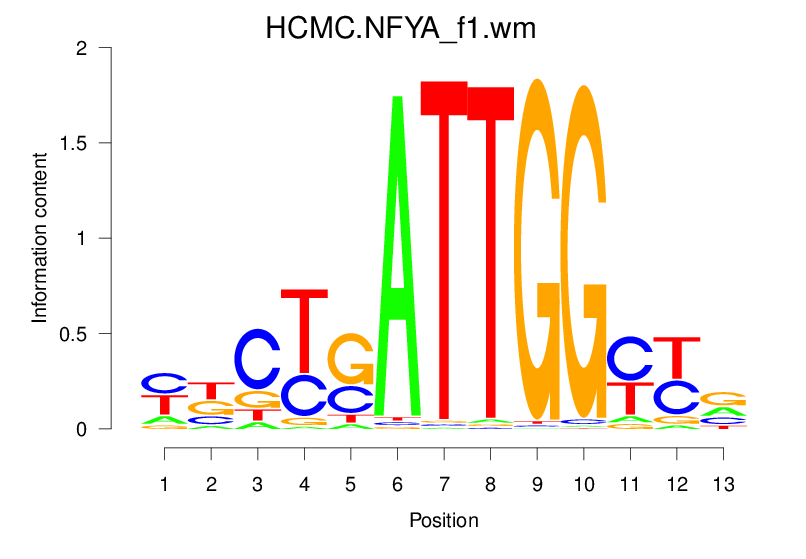 |
10.218 (1162.069) | 2.193 (392.558) | 0.4510 | 0.343 | 4.0781 | 3983/37435 |
| denovo_WM_17 | 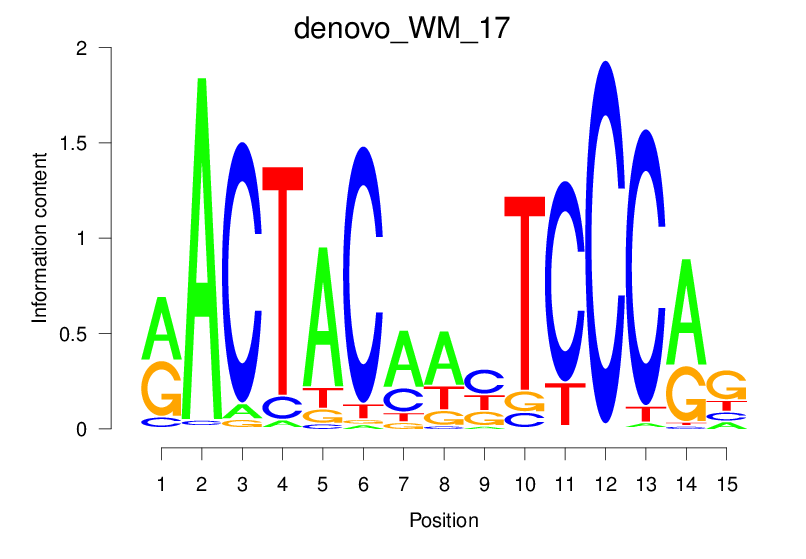 |
11.85 (1236.149) | 1.131 (61.375) | 0.1837 | 0.1478 | 3.3121 | 1577/37435 |
| HTSELEX.JDP2.bZIP.full.dimeric.wm2 | 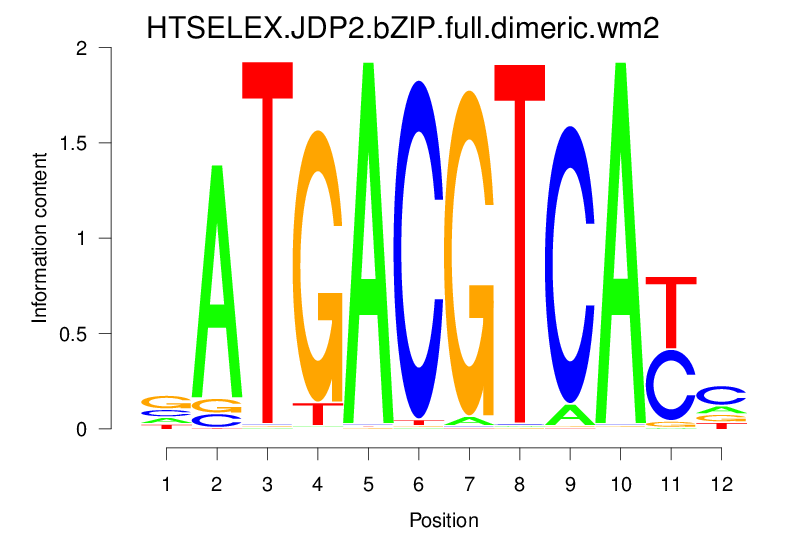 |
12.511 (1263.313) | 1.003 (1.484) | 0.1046 | 0.0441 | 3.3069 | 257/37435 |
| HTSELEX.ETV6.ETS.full.dimeric.wm1 | 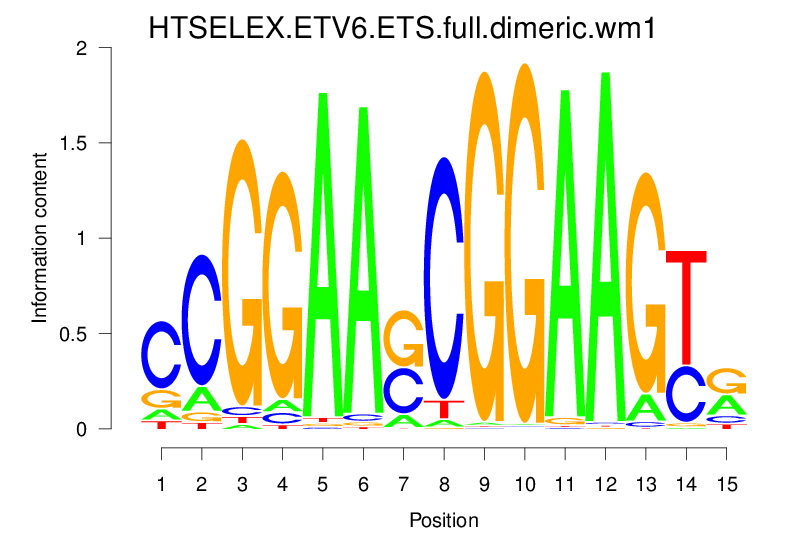 |
13.017 (1283.116) | 1.009 (4.479) | 0.1211 | 0.0373 | 2.7418 | 341/37435 |
| HOMER.RLR1?.SacCer-Promoters | 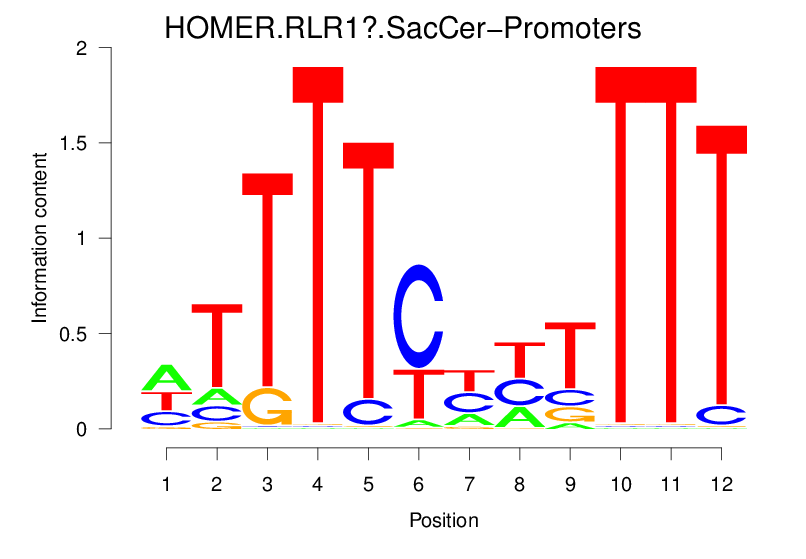 |
13.325 (1294.837) | 1.073 (35.117) | 0.2597 | -0.105 | 0.6335 | 3801/37435 |
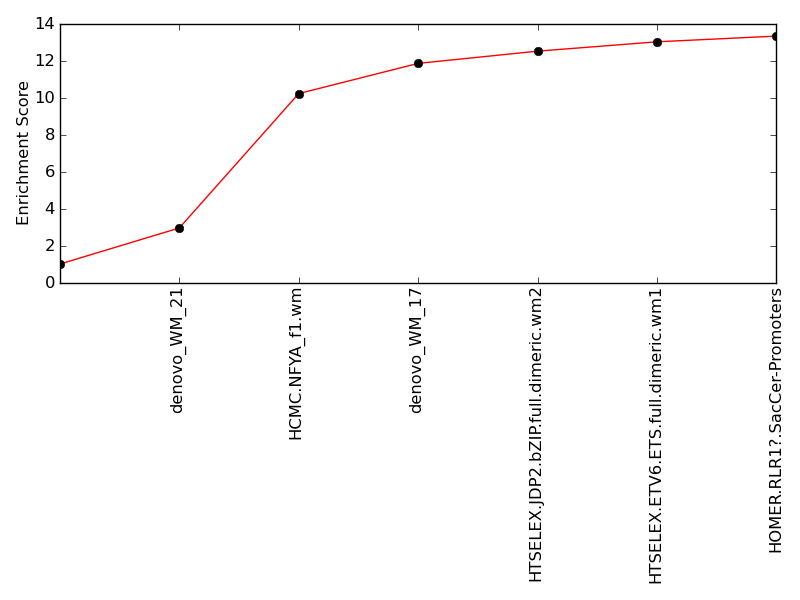
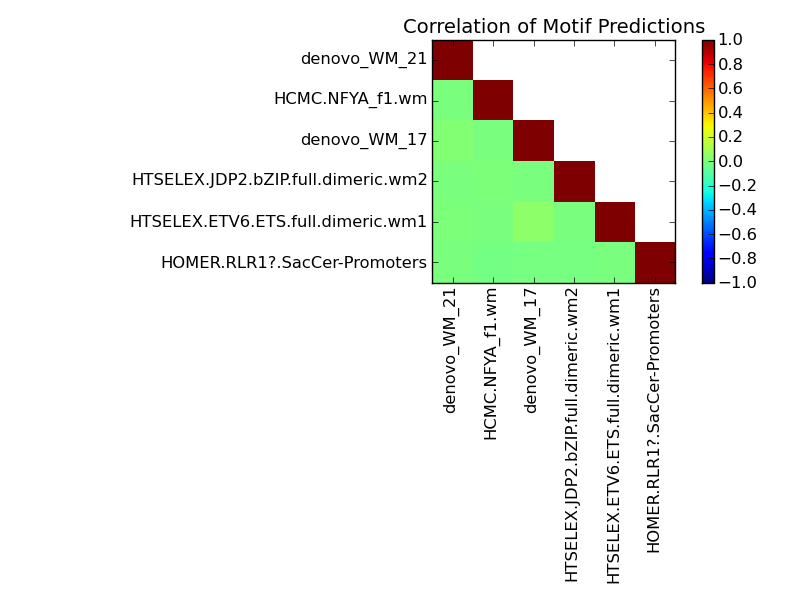
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| denovo_WM_21 |  |
2.951 (541.109) | 0.5226 | 0.4347 | 17.0817 | 1334/37435 |
| denovo_WM_5 | 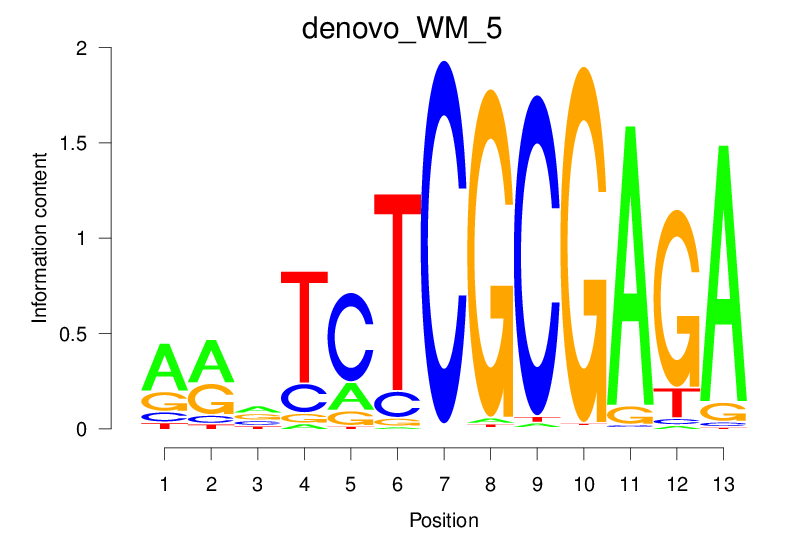 |
2.916 (535.152) | 0.5220 | 0.4322 | 15.864 | 1425/37435 |
| denovo_WM_19 | 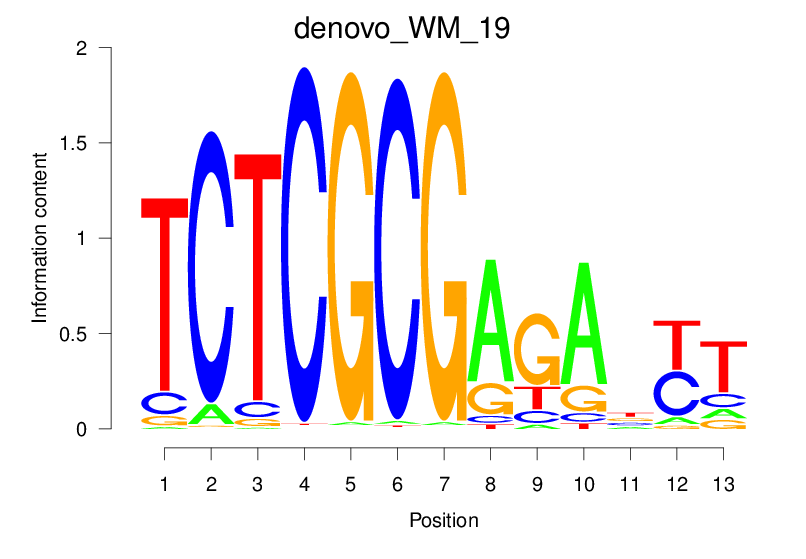 |
2.897 (531.882) | 0.5269 | 0.4298 | 12.8959 | 1832/37435 |
| denovo_WM_13 | 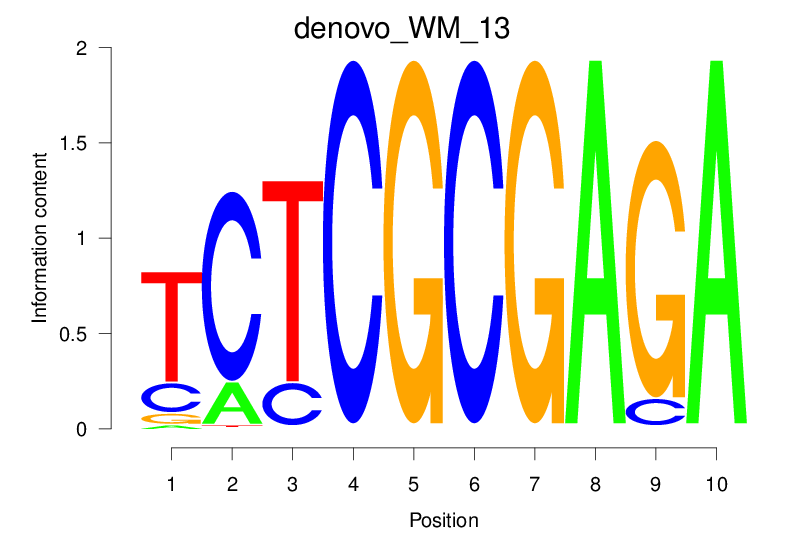 |
2.559 (469.88) | 0.4793 | 0.4054 | 22.7271 | 831/37435 |
| HCMC.NFYA_f1.wm |  |
2.193 (392.558) | 0.4510 | 0.343 | 4.0781 | 3983/37435 |
| HCMC.NFYB_f1.wm | 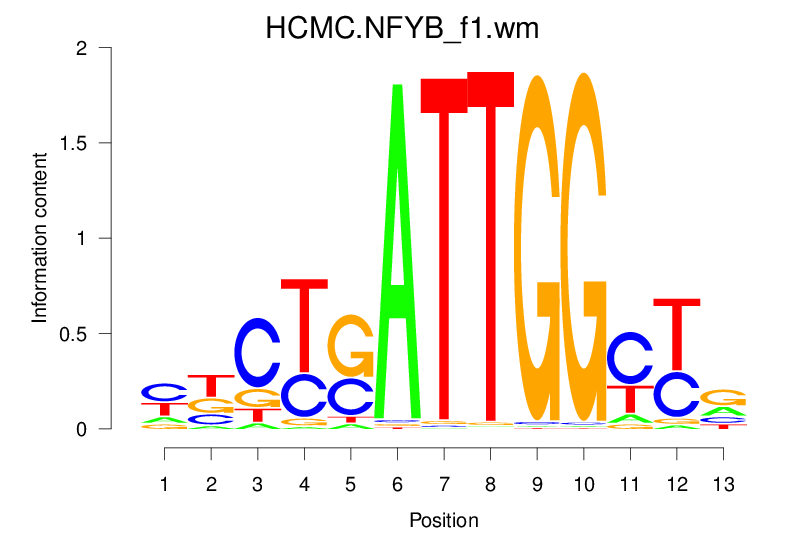 |
2.178 (389.149) | 0.4436 | 0.3342 | 4.1586 | 3763/37435 |
| denovo_WM_7 | 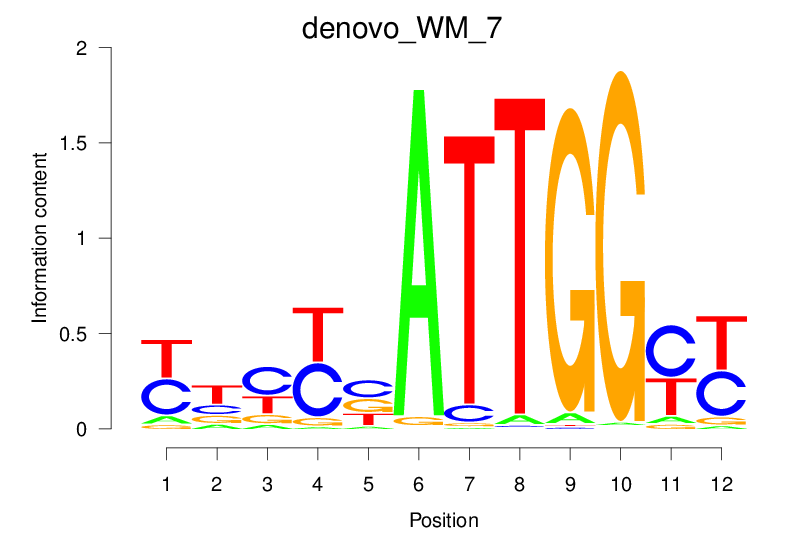 |
2.161 (385.261) | 0.4600 | 0.3455 | 3.7779 | 4091/37435 |
| JASPAR.NFYA.wm | 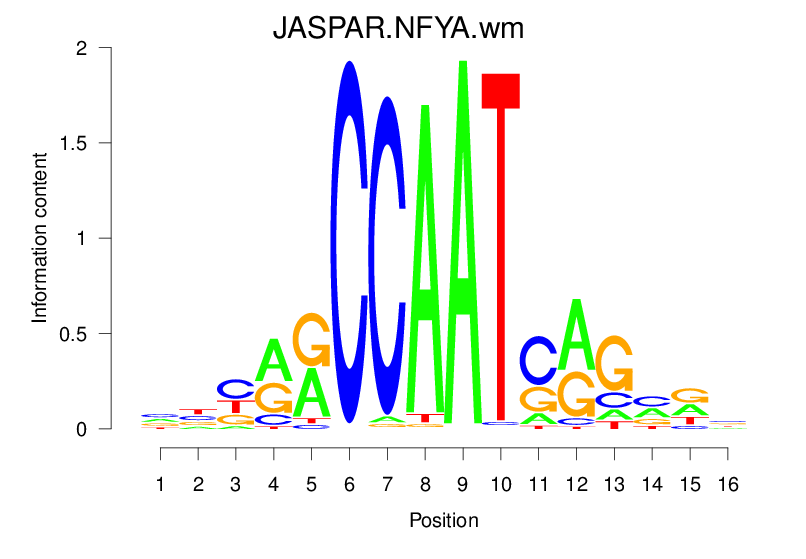 |
2.093 (369.361) | 0.4398 | 0.3477 | 4.0866 | 4122/37435 |
| denovo_WM_8 | 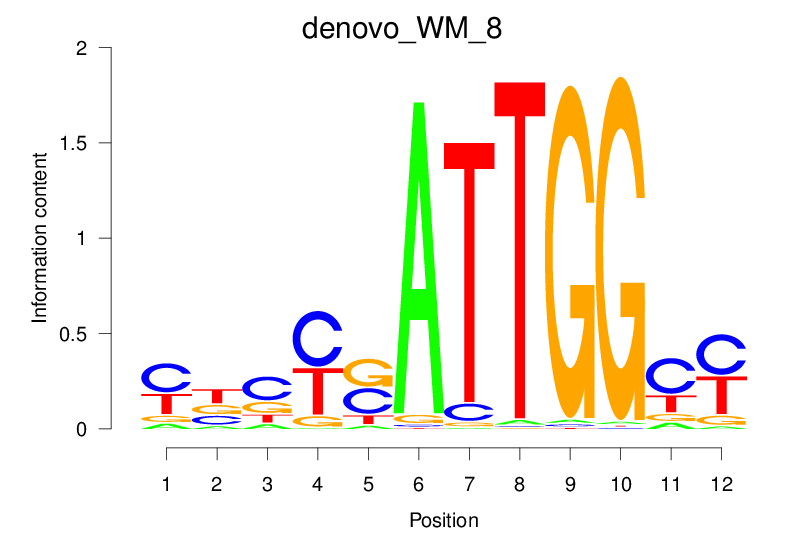 |
2.009 (348.895) | 0.4668 | 0.3646 | 3.368 | 5456/37435 |
| denovo_WM_16 | 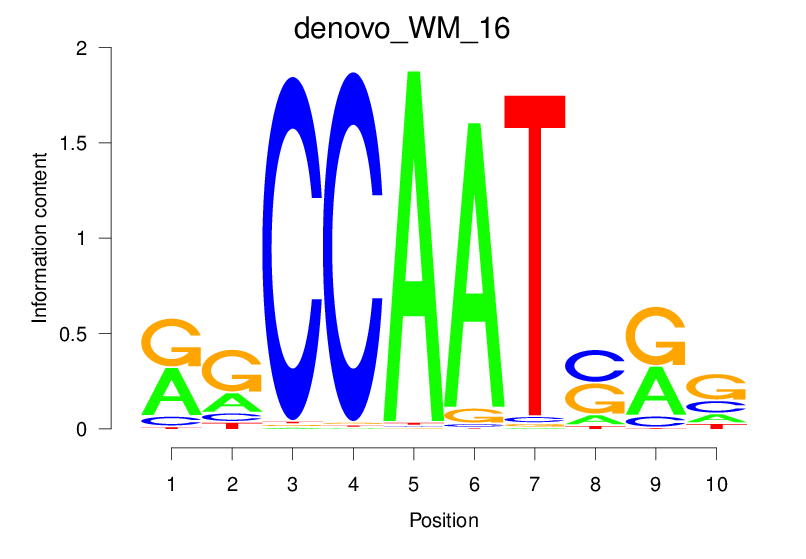 |
1.946 (332.857) | 0.4485 | 0.3556 | 3.3701 | 5224/37435 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis