CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.817 | 65th percentile | |
Fraction of mapped reads for the background samples: |
0.890 | 78th percentile | |
ChIP enrichment signal |
|||
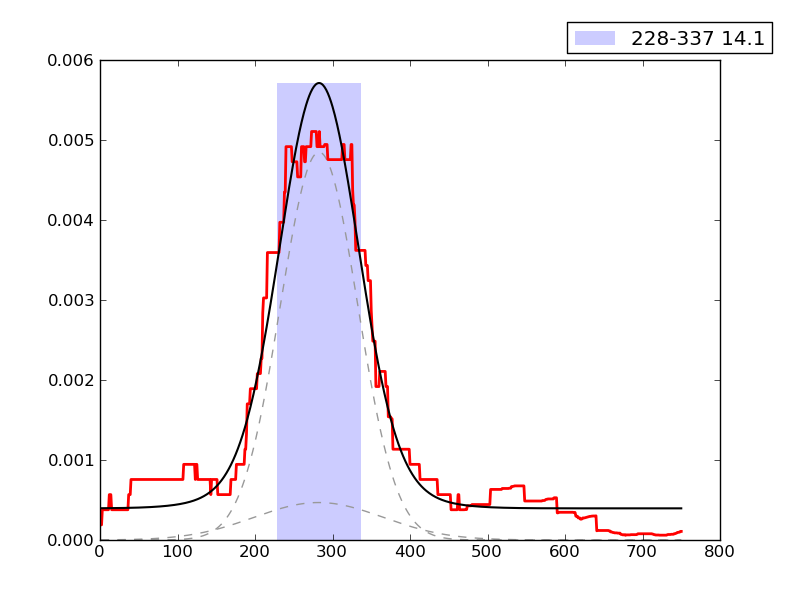
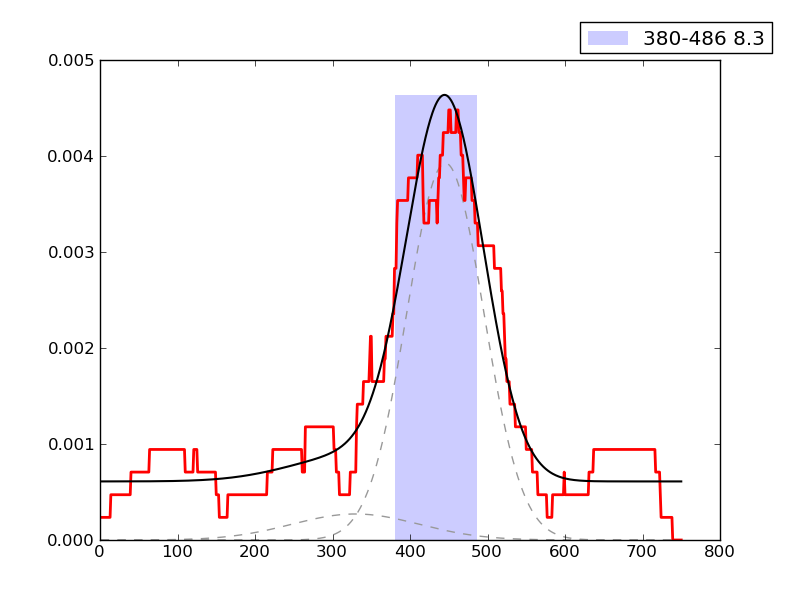
Noise level ChIP signal: |
0.113 | 11th percentile | |
Error in fit of enrichment distribution: |
0.523 | 46th percentile | |
Peak statistics |
|||
Number of peaks: |
1464 | 14th percentile | |
Fraction of reads mapped to peaks: |
0.002 | 8th percentile | |
Motif enrichment statistics |
|||
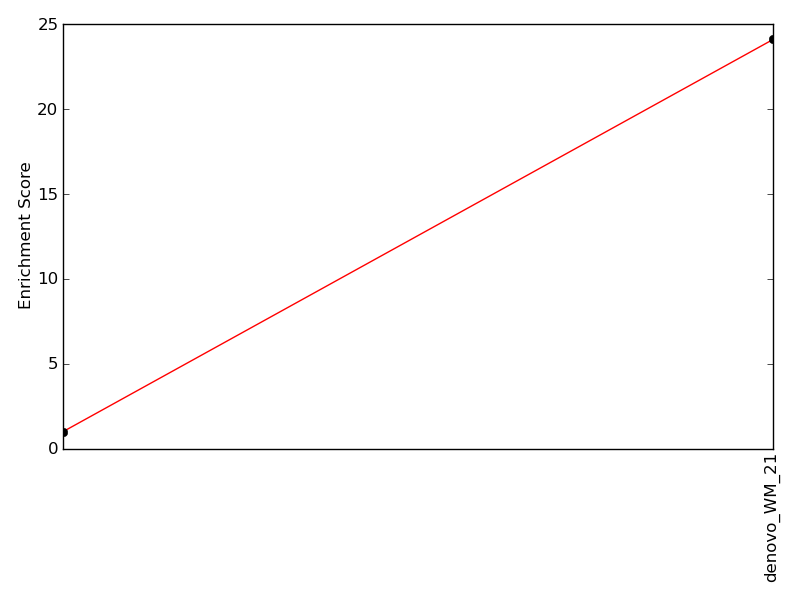
Enrichment of top motif: |
24.11 | 78th percentile | |
Enrichment of complimentary motif set: |
24.11 | 76th percentile | |
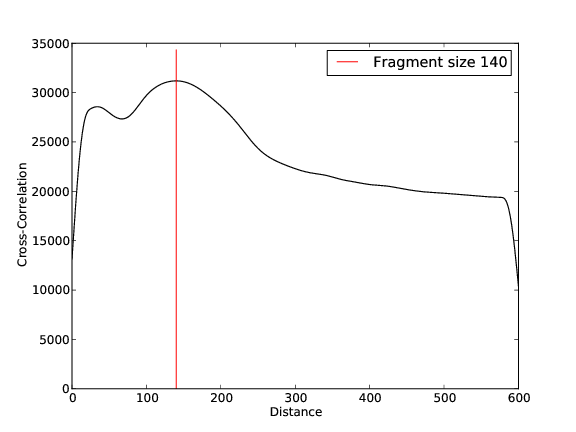
Quality Control, Mapping, Fragment Size Estimation:
IRF3 Replicate 1
- 24,420,258 input reads (in FASTQ format).
- 20,593,022 reads after removal of low quality reads.
- 20,528,656 reads after removal of adapters and low complexity reads.
- 19,148,937 reads after mapping.
- Report PDF
IRF3 Replicate 2
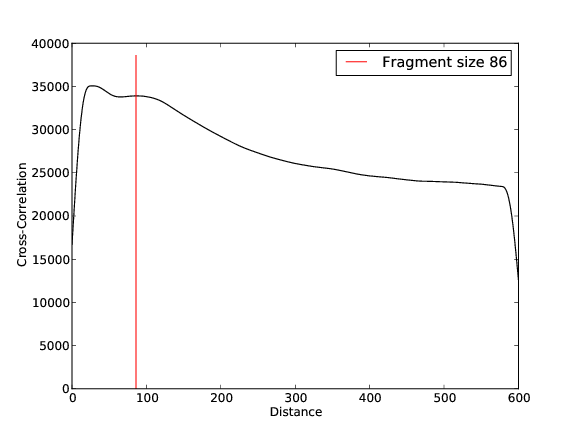
- 30,687,720 input reads (in FASTQ format).
- 27,838,418 reads after removal of low quality reads.
- 27,777,950 reads after removal of adapters and low complexity reads.
- 25,857,877 reads after mapping.
- Report PDF
BG Replicate 1
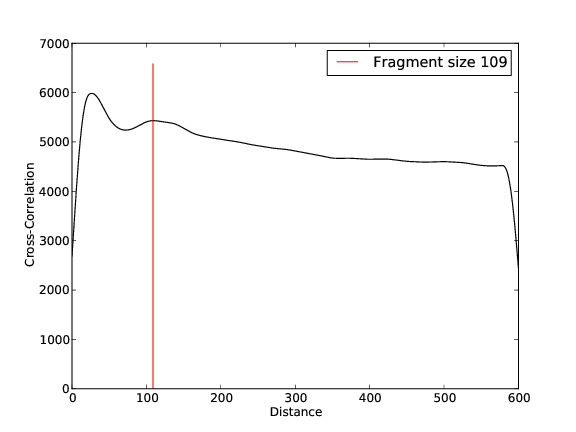
- 6,211,790 input reads (in FASTQ format).
- 6,149,250 reads after removal of low quality reads.
- 6,117,625 reads after removal of adapters and low complexity reads.
- 5,587,889 reads after mapping.
- Report PDF
BG Replicate 2
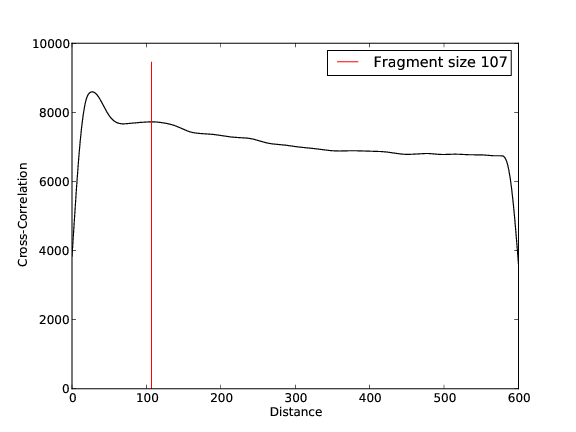
- 5,526,289 input reads (in FASTQ format).
- 5,459,026 reads after removal of low quality reads.
- 5,427,858 reads after removal of adapters and low complexity reads.
- 4,855,998 reads after mapping.
- Report PDF
Peak Calling:
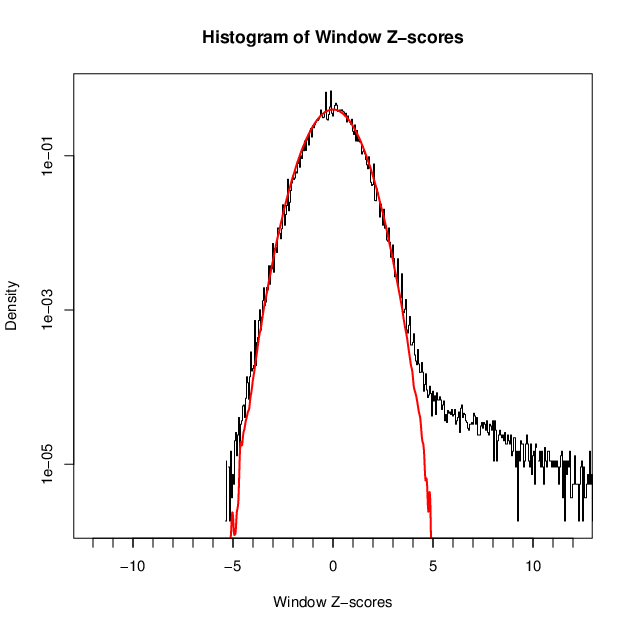
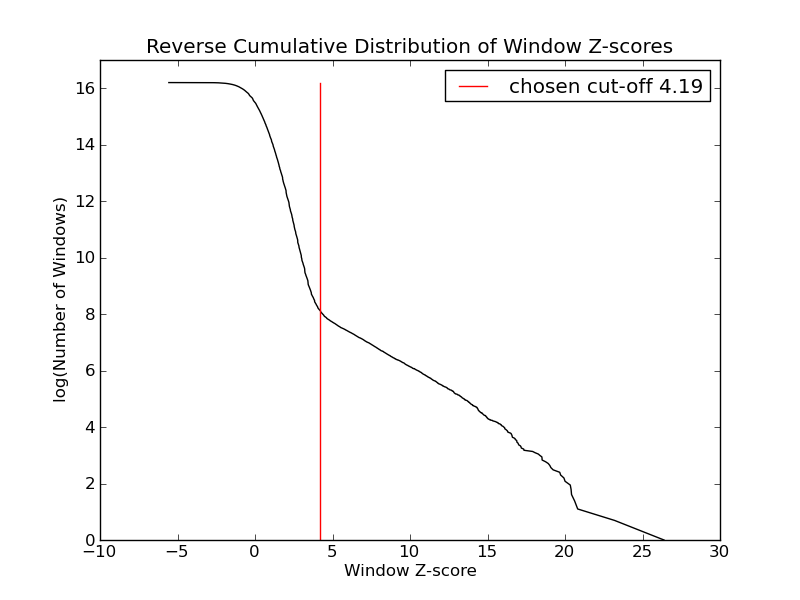
- 3396 windows are significantly enriched at FDR=10% (z-value cut-off: 4.19)
- 3396 windows merged to 1969 regions.
- 1464 significant peaks fitted within 1969 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | denovo_WM_21 |
|---|---|---|---|---|---|---|
| chr21:47604436..47604569 | 22.805 | C21orf56|chromosome 21 open reading frame 56 | -127 (-) | LSS|lanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) | 44003 (-) | |
| chr11:63953450..63953592 | 20.703 | MACROD1|MACRO domain containing 1 | -19966 (-) | STIP1|stress-induced-phosphoprotein 1 | 84 (+) | 1:0.910, 41:0.374 |
| chr17:26368506..26368679 | 19.786 | C17orf108|chromosome 17 open reading frame 108 | -148201 (-) | NLK|nemo-like kinase | 1061 (+) | -28:0.900, -70:0.406 |
| chr11:59383371..59383555 | 19.133 | OSBP|oxysterol binding protein | -253 (-) | OSBP|oxysterol binding protein | 153 (-) | -16:0.238, -36:0.947, 79:0.922 |
| chr7:100184107..100184256 | 18.845 | FBXO24|F-box protein 24 | -179 (+) | FBXO24|F-box protein 24 | 3042 (+) | 2:0.695, -38:0.253 |
| chr17:45973315..45973481 | 18.830 | -248 (-) | SP2|Sp2 transcription factor | 182 (+) | -28:0.657 |
|
| chr14:58765022..58765167 | 18.684 | FLJ31306|uncharacterized LOC379025 | -261 (-) | ARID4A|AT rich interactive domain 4A (RBP1-like) | 147 (+) | 5:0.717, 73:0.708 |
| chr17:17875917..17876098 | 18.440 | TOM1L2|target of myb1-like 2 (chicken) | -266 (-) | LRRC48|leucine rich repeat containing 48 | 193 (+) | |
| chr19:6372587..6372729 | 18.282 | ALKBH7|alkB, alkylation repair homolog 7 (E. coli) | -215 (+) | ALKBH7|alkB, alkylation repair homolog 7 (E. coli) | 96 (+) | |
| chr12:53773609..53773794 | 17.497 | SP7|Sp7 transcription factor | -43698 (-) | SP1|Sp1 transcription factor | 289 (+) | 45:0.635, -89:0.411 |
| chr19:14117230..14117401 | 17.426 | RFX1|regulatory factor X, 1 (influences HLA class II expression) | -186 (-) | RLN3|relaxin 3 | 21672 (+) | |
| chr11:63953199..63953372 | 17.293 | MACROD1|MACRO domain containing 1 | -19730 (-) | STIP1|stress-induced-phosphoprotein 1 | 320 (+) | -54:0.947, 67:0.792 |
| chr9:130829691..130829850 | 17.267 | NAIF1|nuclear apoptosis inducing factor 1 | -172 (-) | SLC25A25|solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 25 | 713 (+) | |
| chr16:30621709..30621839 | 17.194 | ZNF689|zinc finger protein 689 | -118 (-) | PRR14|proline rich 14 | 40326 (+) | -10:0.266, 20:0.346 |
| chr17:61627591..61627774 | 17.178 | KCNH6|potassium voltage-gated channel, subfamily H (eag-related), member 6 | -26958 (+) | DCAF7|DDB1 and CUL4 associated factor 7 | 144 (+) | 36:0.846 |
| chr11:67275695..67275840 | 17.008 | CDK2AP2|cyclin-dependent kinase 2 associated protein 2 | -134 (-) | CDK2AP2|cyclin-dependent kinase 2 associated protein 2 | 372 (-) | 18:0.856, -21:0.799 |
| chr17:38574229..38574395 | 16.975 | TOP2A|topoisomerase (DNA) II alpha 170kDa | -188 (-) | IGFBP4|insulin-like growth factor binding protein 4 | 25507 (+) | -4:0.499, -71:0.442, 79:0.972 |
| chr19:7894814..7894976 | 16.961 | CLEC4M|C-type lectin domain family 4, member M | -66802 (+) | EVI5L|ecotropic viral integration site 5-like | 281 (+) | -43:0.521 |
| chr19:12662360..12662504 | 16.890 | ZNF564|zinc finger protein 564 | -159 (-) | ZNF490|zinc finger protein 490 | 59129 (-) | -86:0.826 |
| chr19:54641214..54641398 | 16.871 | PRPF31|PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) | -22127 (+) | CNOT3|CCR4-NOT transcription complex, subunit 3 | 141 (+) | 47:0.755 |
| chr17:79818616..79818771 | 16.750 | P4HB|prolyl 4-hydroxylase, beta polypeptide | -156 (-) | ARHGDIA|Rho GDP dissociation inhibitor (GDI) alpha | 10047 (-) | -17:0.988, 98:0.893 |
| chr17:4870610..4870782 | 16.658 | ENO3|enolase 3 (beta, muscle) | -16292 (+) | SPAG7|sperm associated antigen 7 | 415 (-) | -31:0.746, -71:0.370 |
| chr19:13905997..13906148 | 16.499 | C19orf53|chromosome 19 open reading frame 53 | -20788 (+) | ZSWIM4|zinc finger, SWIM-type containing 4 | 173 (+) | -42:0.988, 87:0.340 |
| chr19:36193319..36193457 | 16.496 | UPK1A|uroplakin 1A | -35672 (+) | ZBTB32|zinc finger and BTB domain containing 32 | 10441 (+) | -22:0.986, 33:0.242, 94:0.966 |
| chr19:47551913..47552033 | 16.443 | TMEM160|transmembrane protein 160 | -77 (-) | ZC3H4|zinc finger CCCH-type containing 4 | 63513 (-) | -10:0.573, 31:0.223 |
| chr9:140135525..140135676 | 16.392 | SLC34A3|solute carrier family 34 (sodium phosphate), member 3 | -10216 (+) | TUBB4B|tubulin, beta 4B class IVb | 108 (+) | -23:0.306 |
| chr8:145159238..145159356 | 16.212 | SHARPIN|SHANK-associated RH domain interactor | -146 (-) | MAF1|MAF1 homolog (S. cerevisiae) | 47 (+) | -77:0.558 |
| chr12:57039840..57039982 | 16.146 | ATP5B|ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide | -85 (-) | PTGES3|prostaglandin E synthase 3 (cytosolic) | 41939 (-) | 40:0.626 |
| chr19:42772981..42773165 | 16.104 | ERF|Ets2 repressor factor | -13768 (-) | CIC|capicua homolog (Drosophila) | 15656 (+) | -42:0.540, -82:0.266 |
| chr19:10828452..10828636 | 16.037 | QTRT1|queuine tRNA-ribosyltransferase 1 | -16386 (+) | DNM2|dynamin 2 | 188 (+) | 0:0.325, 100:0.921 |
| chr19:4342840..4342997 | 15.999 | STAP2|signal transducing adaptor family member 2 | -168 (-) | MPND|MPN domain containing | 641 (+) | -27:0.620, 49:0.227, -51:0.798, 83:0.564 |
| chr14:96001104..96001269 | 15.964 | SNHG10|small nucleolar RNA host gene 10 (non-protein coding) | -171 (-) | GLRX5|glutaredoxin 5 | 140 (+) | 109:0.257 |
| chr19:15529839..15529998 | 15.861 | AKAP8L|A kinase (PRKA) anchor protein 8-like | -84 (-) | WIZ|widely interspaced zinc finger motifs | 13627 (-) | 87:0.312 |
| chr1:206138208..206138393 | 15.859 | LOC100132966|uncharacterized LOC100132966 | -852 (-) | FAM72A|family with sequence similarity 72, member A FAM72B|family with sequence similarity 72, member B | 616 (+) | -16:0.582, -47:0.770 |
| chr1:120838743..120838928 | 15.847 | NOTCH2|notch 2 | -226520 (-) | FAM72B|family with sequence similarity 72, member B | 169 (+) | -54:0.770 |
| chr19:6424941..6425116 | 15.840 | KHSRP|KH-type splicing regulatory protein | -206 (-) | SLC25A41|solute carrier family 25, member 41 | 8752 (-) | -63:0.971 |
| chr19:42773250..42773375 | 15.825 | ERF|Ets2 repressor factor | -14007 (-) | CIC|capicua homolog (Drosophila) | 15417 (+) | 57:0.966 |
| chr19:6425242..6425424 | 15.713 | KHSRP|KH-type splicing regulatory protein | -511 (-) | SLC25A41|solute carrier family 25, member 41 | 8447 (-) | -94:0.397 |
| chr17:79212906..79213057 | 15.663 | C17orf56|chromosome 17 open reading frame 56 | -92 (-) | C17orf89|chromosome 17 open reading frame 89 | 153 (+) | -5:0.498 |
| chr1:143913327..143913512 | 15.612 | FAM72D|family with sequence similarity 72, member D | -289 (-) | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | 450805 (-) | 48:0.468, -71:0.770 |
| chr7:106809257..106809398 | 15.602 | PRKAR2B|protein kinase, cAMP-dependent, regulatory, type II, beta | -124124 (+) | HBP1|HMG-box transcription factor 1 | 112 (+) | 49:0.918, -94:0.222 |
| chr9:139780736..139780861 | 15.565 | EDF1|endothelial differentiation-related factor 1 | -20060 (-) | TRAF2|TNF receptor-associated factor 2 | 155 (+) | -4:0.221, -38:0.789 |
| chr14:21737674..21737818 | 15.504 | HNRNPC|heterogeneous nuclear ribonucleoprotein C (C1/C2) | -175 (-) | RPGRIP1|retinitis pigmentosa GTPase regulator interacting protein 1 | 18389 (+) | -25:0.830, 82:0.920, -93:0.965 |
| chr1:151763080..151763252 | 15.419 | TDRKH|tudor and KH domain containing | -196 (-) | 98 (+) | 10:0.862, 73:0.324 |
|
| chr19:6737680..6737831 | 15.371 | GPR108|G protein-coupled receptor 108 | -137 (-) | TRIP10|thyroid hormone receptor interactor 10 | 1979 (+) | -54:0.938, -104:0.593 |
| chr19:10828308..10828427 | 15.307 | QTRT1|queuine tRNA-ribosyltransferase 1 | -16209 (+) | DNM2|dynamin 2 | 365 (+) | 1:0.949 |
| chr19:37808566..37808727 | 15.189 | ZNF383|zinc finger protein 383 | -91281 (+) | HKR1|HKR1, GLI-Kruppel zinc finger family member | 179 (+) | -23:0.985, 111:0.904 |
| chr14:104181817..104181948 | 14.983 | XRCC3|X-ray repair complementing defective repair in Chinese hamster cells 3 | -99 (-) | ZFYVE21|zinc finger, FYVE domain containing 21 | 234 (+) | -28:0.685 |
| chr15:89631446..89631607 | 14.957 | ABHD2|abhydrolase domain containing 2 | -146 (+) | ABHD2|abhydrolase domain containing 2 | 122 (+) | |
| chr20:32308110..32308260 | 14.868 | PXMP4|peroxisomal membrane protein 4, 24kDa | -171 (-) | ZNF341|zinc finger protein 341 | 11611 (+) | -23:0.981, 60:0.952 |
| chr2:27008668..27008825 | 14.864 | C2orf18|chromosome 2 open reading frame 18 CENPA|centromere protein A | -21577 (+) | CENPA|centromere protein A | 146 (+) | -26:0.349, 48:0.976 |
| chr19:2945001..2945107 | 14.786 | ZNF77|zinc finger protein 77 | -111 (-) | TLE6|transducin-like enhancer of split 6 (E(sp1) homolog, Drosophila) | 32512 (+) | -35:0.959, 68:0.959, 106:0.826 |
| chr2:88927129..88927302 | 14.703 | EIF2AK3|eukaryotic translation initiation factor 2-alpha kinase 3 | -237 (-) | RPIA|ribose 5-phosphate isomerase A | 64000 (+) | |
| chr7:44887893..44888034 | 14.609 | H2AFV|H2A histone family, member V | -237 (-) | PURB|purine-rich element binding protein B | 37006 (-) | -14:0.921, -45:0.809 |
| chr17:74667855..74668000 | 14.377 | ST6GALNAC1|ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 1 | -28121 (-) | MXRA7|matrix-remodelling associated 7 | 39105 (-) | 60:0.364 |
| chr5:139781055..139781217 | 14.353 | -95 (-) | ANKHD1-EIF4EBP3|ANKHD1-EIF4EBP3 readthrough ANKHD1|ankyrin repeat and KH domain containing 1 | 261 (+) | 63:0.298 |
|
| chr5:151151258..151151443 | 14.276 | LOC100652758|uncharacterized LOC100652758 | -377 (-) | G3BP1|GTPase activating protein (SH3 domain) binding protein 1 | 122 (+) | 11:0.744, -63:0.344 |
| chr12:56367552..56367737 | 14.247 | CDK2|cyclin-dependent kinase 2 | -7055 (+) | RAB5B|RAB5B, member RAS oncogene family | 118 (+) | 21:0.291 |
| chr19:4246936..4247073 | 14.149 | EBI3|Epstein-Barr virus induced 3 | -17484 (+) | CCDC94|coiled-coil domain containing 94 | 86 (+) | -6:0.902, 25:0.587 |
| chr19:4968723..4968907 | 14.115 | -51987 (-) | KDM4B|lysine (K)-specific demethylase 4B | 309 (+) | 44:0.301 |
|
| chr19:4472050..4472177 | 14.113 | UBXN6|UBX domain protein 6 | -14324 (-) | HDGFRP2|hepatoma-derived growth factor-related protein 2 | 234 (+) | -22:0.954 |
| chr19:42901131..42901279 | 14.061 | CNFN|cornifelin | -6774 (-) | 107 (+) | -49:0.475 |
|
| chr22:40440627..40440763 | 13.988 | FAM83F|family with sequence similarity 83, member F | -49751 (+) | TNRC6B|trinucleotide repeat containing 6B | 214 (+) | -47:0.862 |
| chr19:17377946..17378103 | 13.914 | USHBP1|Usher syndrome 1C binding protein 1 | -2477 (-) | BABAM1|BRISC and BRCA1 A complex member 1 | 181 (+) | 21:0.317, 53:0.618 |
| chr1:43312112..43312233 | 13.860 | ERMAP|erythroblast membrane-associated protein (Scianna blood group) | -20902 (+) | ZNF691|zinc finger protein 691 | 78 (+) | 22:0.839 |
| chr12:53773362..53773546 | 13.811 | SP7|Sp7 transcription factor | -43451 (-) | SP1|Sp1 transcription factor | 536 (+) | 66:0.989, -104:0.740 |
| chr22:38054356..38054512 | 13.807 | SH3BP1|SH3-domain binding protein 1 | -18675 (+) | PDXP|pyridoxal (pyridoxine, vitamin B6) phosphatase | 311 (+) | 7:0.977, -44:0.993 |
| chr1:6845206..6845359 | 13.783 | DNAJC11|DnaJ (Hsp40) homolog, subfamily C, member 11 | -83351 (-) | CAMTA1|calmodulin binding transcription activator 1 | 209 (+) | -49:0.705 |
| chr1:6052577..6052705 | 13.778 | NPHP4|nephronophthisis 4 | -132 (-) | KCNAB2|potassium voltage-gated channel, shaker-related subfamily, beta member 2 | 85 (+) | 15:0.256, -38:0.755, 46:0.993, -81:0.994 |
| chr1:151254512..151254677 | 13.713 | ZNF687|zinc finger protein 687 | -552 (+) | ZNF687|zinc finger protein 687 | 179 (+) | -47:0.781 |
| chr9:37465687..37465872 | 13.679 | ZBTB5|zinc finger and BTB domain containing 5 | -338 (-) | POLR1E|polymerase (RNA) I polypeptide E, 53kDa | 20251 (+) | -3:0.939, 47:0.266 |
| chr19:18315084..18315200 | 13.672 | RAB3A|RAB3A, member RAS oncogene family | -338 (-) | PDE4C|phosphodiesterase 4C, cAMP-specific | 20160 (-) | -2:0.994 |
| chr2:73460425..73460544 | 13.656 | PRADC1|protease-associated domain containing 1 | -120 (-) | CCT7|chaperonin containing TCP1, subunit 7 (eta) | 915 (+) | 18:0.898 |
| chr12:14927151..14927276 | 13.555 | HIST4H4|histone cluster 4, H4 | -3172 (-) | H2AFJ|H2A histone family, member J | 72 (+) | 47:0.763, 64:0.839 |
| chr19:18303745..18303929 | 13.514 | IFI30|interferon, gamma-inducible protein 30 | -19196 (+) | MPV17L2|MPV17 mitochondrial membrane protein-like 2 | 198 (+) | -21:0.949, 41:0.812 |
| chr19:19174380..19174540 | 13.485 | ARMC6|armadillo repeat containing 6 | -29800 (+) | SLC25A42|solute carrier family 25, member 42 | 349 (+) | |
| chr9:131843085..131843265 | 13.474 | FAM73B|family with sequence similarity 73, member B | -43943 (+) | DOLPP1|dolichyl pyrophosphate phosphatase 1 | 220 (+) | -11:0.988, -59:0.993, 71:0.329 |
| chr2:71454254..71454403 | 13.449 | PAIP2B|poly(A) binding protein interacting protein 2B | -98 (-) | ZNF638|zinc finger protein 638 | 49398 (+) | 19:0.786, 74:0.989 |
| chr22:35696129..35696255 | 13.410 | TOM1|target of myb1 (chicken) | -296 (+) | HMOX1|heme oxygenase (decycling) 1 | 80876 (+) | |
| chr19:33462937..33463061 | 13.401 | CEP89|centrosomal protein 89kDa | -156 (-) | C19orf40|chromosome 19 open reading frame 40 | 171 (+) | -50:0.981, 50:0.961, -75:0.426 |
| chr7:27702580..27702717 | 13.399 | HIBADH|3-hydroxyisobutyrate dehydrogenase | -71 (-) | TAX1BP1|Tax1 (human T-cell leukemia virus type I) binding protein 1 | 76343 (+) | -8:0.978, 28:0.961 |
| chr5:145826719..145826864 | 13.349 | POU4F3|POU class 4 homeobox 3 | -108161 (+) | TCERG1|transcription elongation regulator 1 | 103 (+) | 77:0.970 |
| chr11:66056638..66056821 | 13.335 | YIF1A|Yip1 interacting factor homolog A (S. cerevisiae) | -105 (-) | TMEM151A|transmembrane protein 151A | 2609 (+) | 73:0.851, -97:0.811 |
| chr20:44486078..44486182 | 13.306 | ACOT8|acyl-CoA thioesterase 8 | -121 (-) | ZSWIM3|zinc finger, SWIM-type containing 3 | 110 (+) | -36:0.446 |
| chr19:48972330..48972509 | 13.271 | KCNJ14|potassium inwardly-rectifying channel, subfamily J, member 14 | -7914 (+) | CYTH2|cytohesin 2 | 99 (+) | 12:0.972, -58:0.301 |
| chr1:154955675..154955805 | 13.210 | CKS1B|CDC28 protein kinase regulatory subunit 1B | -8509 (+) | FLAD1|FAD1 flavin adenine dinucleotide synthetase homolog (S. cerevisiae) | 63 (+) | |
| chr12:54718751..54718892 | 13.210 | NFE2|nuclear factor (erythroid-derived 2), 45kDa | -24014 (-) | COPZ1|coatomer protein complex, subunit zeta 1 | 91 (+) | 73:0.934 |
| chr9:34989471..34989655 | 13.206 | KIAA1045|KIAA1045 | -31240 (+) | DNAJB5|DnaJ (Hsp40) homolog, subfamily B, member 5 | 154 (+) | -41:0.218 |
| chr11:125462549..125462699 | 13.200 | EI24|etoposide induced 2.4 mRNA | -23217 (+) | STT3A|STT3, subunit of the oligosaccharyltransferase complex, homolog A (S. cerevisiae) | 114 (+) | |
| chr16:1832631..1832815 | 13.159 | SPSB3|splA/ryanodine receptor domain and SOCS box containing 3 | -131 (-) | NUBP2|nucleotide binding protein 2 | 236 (+) | -6:0.898 |
| chr19:1041053..1041190 | 13.154 | ABCA7|ATP-binding cassette, sub-family A (ABC1), member 7 | -987 (+) | HMHA1|histocompatibility (minor) HA-1 | 26022 (+) | -2:0.929, 31:0.956 |
| chr16:57769234..57769374 | 13.104 | CCDC135|coiled-coil domain containing 135 | -40575 (+) | KATNB1|katanin p80 (WD repeat containing) subunit B 1 | 341 (+) | 4:0.899, -38:0.967 |
| chr19:36248846..36248962 | 13.085 | HSPB6|heat shock protein, alpha-crystallin-related, B6 | -973 (-) | C19orf55|chromosome 19 open reading frame 55 | 173 (+) | -60:0.285, 70:0.821 |
| chr1:110881830..110882015 | 12.955 | KCNC4|potassium voltage-gated channel, Shaw-related subfamily, member 4 | -127925 (+) | RBM15|RNA binding motif protein 15 | 72 (+) | -26:0.337, 106:0.809 |
| chr19:14228600..14228721 | 12.945 | PRKACA|protein kinase, cAMP-dependent, catalytic, alpha | -91 (-) | ASF1B|ASF1 anti-silencing function 1 homolog B (S. cerevisiae) | 18672 (-) | -18:0.946, -61:0.878 |
| chr19:50269732..50269916 | 12.887 | TSKS|testis-specific serine kinase substrate | -3296 (-) | AP2A1|adaptor-related protein complex 2, alpha 1 subunit | 327 (+) | 44:0.444 |
| chr19:45873883..45874006 | 12.844 | ERCC2|excision repair cross-complementing rodent repair deficiency, complementation group 2 | -99 (-) | PPP1R13L|protein phosphatase 1, regulatory subunit 13 like | 34369 (-) | 4:0.875, 97:0.924 |
| chr14:94492590..94492686 | 12.843 | ASB2|ankyrin repeat and SOCS box containing 2 | -49562 (-) | OTUB2|OTU domain, ubiquitin aldehyde binding 2 | 65 (+) | |
| chr4:52709021..52709158 | 12.827 | CWH43|cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) | -3720817 (+) | DCUN1D4|DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) | 197 (+) | |
| chrX:49047673..49047857 | 12.827 | PRICKLE3|prickle homolog 3 (Drosophila) | -4911 (-) | 125 (+) | -13:0.744, 57:0.980 |
|
| chr19:41769874..41770056 | 12.822 | HNRNPUL1|heterogeneous nuclear ribonucleoprotein U-like 1 | -1498 (+) | HNRNPUL1|heterogeneous nuclear ribonucleoprotein U-like 1 | 184 (+) | 8:0.415, 38:0.641, -80:0.439 |
| chr3:88108203..88108318 | 12.820 | CGGBP1|CGG triplet repeat binding protein 1 | -117 (-) | ZNF654|zinc finger protein 654 | 80001 (+) | |
| chr19:39340661..39340845 | 12.807 | HNRNPL|heterogeneous nuclear ribonucleoprotein L | -128 (-) | HNRNPL|heterogeneous nuclear ribonucleoprotein L | 2223 (-) | -9:0.659 |
| chrX:49125622..49125807 | 12.772 | FOXP3|forkhead box P3 | -4485 (-) | PPP1R3F|protein phosphatase 1, regulatory subunit 3F | 589 (+) | -25:0.423, 25:0.486, -77:0.908, 88:0.781 |
| chr1:206137850..206138035 | 12.731 | LOC100132966|uncharacterized LOC100132966 | -494 (-) | FAM72A|family with sequence similarity 72, member A FAM72B|family with sequence similarity 72, member B | 974 (+) | -50:0.965 |
| chr2:97523816..97523943 | 12.731 | ANKRD39|ankyrin repeat domain 39 | -137 (-) | SEMA4C|sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4C | 11752 (-) | |
| chr1:150601634..150601792 | 12.716 | ENSA|endosulfine alpha | -131 (-) | ENSA|endosulfine alpha | 216 (-) | -30:0.891 |
| chr14:23755389..23755526 | 12.620 | HOMEZ|homeobox and leucine zipper encoding | -132 (-) | BCL2L2-PABPN1|BCL2L2-PABPN1 readthrough BCL2L2|BCL2-like 2 | 20587 (+) | -18:0.388 |
| chr2:44395513..44395689 | 12.616 | LRPPRC|leucine-rich PPR-motif containing | -172462 (-) | PPM1B|protein phosphatase, Mg2+/Mn2+ dependent, 1B | 414 (+) | -40:0.954 |
| chr11:13299045..13299189 | 12.589 | RASSF10|Ras association (RalGDS/AF-6) domain family (N-terminal) member 10 | -268193 (+) | ARNTL|aryl hydrocarbon receptor nuclear translocator-like | 175 (+) | 35:0.287, 81:0.664 |
| chr3:126422969..126423075 | 12.571 | TXNRD3|thioredoxin reductase 3 | -49046 (-) | CHCHD6|coiled-coil-helix-coiled-coil-helix domain containing 6 | 55 (+) | |
| chr19:55851070..55851208 | 12.520 | TMEM150B|transmembrane protein 150B | -14450 (-) | SUV420H2|suppressor of variegation 4-20 homolog 2 (Drosophila) | 121 (+) | -8:0.946, 44:0.710 |
| chr16:74700813..74700972 | 12.513 | RFWD3|ring finger and WD repeat domain 3 | -115 (-) | MLKL|mixed lineage kinase domain-like | 33845 (-) | 18:0.988, -64:0.824 |
| chr19:54663563..54663741 | 12.500 | LENG1|leukocyte receptor cluster (LRC) member 1 | -194 (-) | TMC4|transmembrane channel-like 4 | 13226 (-) | 19:0.616, 63:0.510, -70:0.219 |
| chr16:89557178..89557337 | 12.492 | ANKRD11|ankyrin repeat domain 11 | -326 (-) | SPG7|spastic paraplegia 7 (pure and complicated autosomal recessive) | 17557 (+) | |
| chr17:57232597..57232775 | 12.465 | SKA2|spindle and kinetochore associated complex subunit 2 | -140 (-) | SKA2|spindle and kinetochore associated complex subunit 2 | 113 (-) | -56:0.789, 77:0.839 |
| chr19:55973067..55973242 | 12.441 | ISOC2|isochorismatase domain containing 2 | -99 (-) | ZNF628|zinc finger protein 628 | 14544 (+) | -78:0.203 |
| chr19:17420202..17420342 | 12.417 | MRPL34|mitochondrial ribosomal protein L34 | -3681 (+) | DDA1|DET1 and DDB1 associated 1 | 9 (+) | 0:0.754, -29:0.792 |
| chr17:73452571..73452700 | 12.383 | GRB2|growth factor receptor-bound protein 2 | -50832 (-) | KIAA0195|KIAA0195 | 103 (+) | -19:0.985, 20:0.495 |
| chr1:150669761..150669920 | 12.363 | GOLPH3L|golgi phosphoprotein 3-like | -255 (-) | HORMAD1|HORMA domain containing 1 | 23502 (-) | -16:0.333, 47:0.966 |
| chr22:20004397..20004514 | 12.355 | ARVCF|armadillo repeat gene deleted in velocardiofacial syndrome | -132 (-) | C22orf25|chromosome 22 open reading frame 25 | 4162 (+) | -46:0.904 |
| chr8:42010303..42010433 | 12.354 | KAT6A|K(lysine) acetyltransferase 6A | -100869 (-) | AP3M2|adaptor-related protein complex 3, mu 2 subunit | 165 (+) | 24:0.804 |
| chr3:114866146..114866331 | 12.325 | ZBTB20|zinc finger and BTB domain containing 20 | -124 (-) | GAP43|growth associated protein 43 | 475912 (+) | -14:0.986 |
| chr4:331425..331558 | 12.296 | ZNF732|zinc finger protein 732 | -41548 (-) | ZNF141|zinc finger protein 141 | 104 (+) | 34:0.890 |
| chr6:136571472..136571636 | 12.291 | FAM54A|family with sequence similarity 54, member A | -125 (-) | BCLAF1|BCL2-associated transcription factor 1 | 39356 (-) | -16:0.978, 64:0.970 |
| chr2:232645999..232646133 | 12.248 | PDE6D|phosphodiesterase 6D, cGMP-specific, rod, delta | -141 (-) | COPS7B|COP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) | 371 (+) | -29:0.767 |
| chr20:17662929..17663062 | 12.242 | RRBP1|ribosome binding protein 1 homolog 180kDa (dog) | -112 (-) | BANF2|barrier to autointegration factor 2 | 11324 (+) | -15:0.904, 90:0.946 |
| chr1:120838359..120838543 | 12.225 | NOTCH2|notch 2 | -226136 (-) | FAM72B|family with sequence similarity 72, member B | 553 (+) | -30:0.965 |
| chr16:30381966..30382151 | 12.181 | TBC1D10B|TBC1 domain family, member 10B | -534 (-) | MYLPF|myosin light chain, phosphorylatable, fast skeletal muscle | 4052 (+) | 1:0.755, 33:0.220, -47:0.981 |
| chr15:90895365..90895490 | 12.175 | NGRN|neugrin, neurite outgrowth associated | -86507 (+) | ZNF774|zinc finger protein 774 | 96 (+) | 28:0.774 |
| chr19:39340963..39341147 | 12.175 | HNRNPL|heterogeneous nuclear ribonucleoprotein L | -430 (-) | HNRNPL|heterogeneous nuclear ribonucleoprotein L | 1921 (-) | 51:0.699, -90:0.821, 113:0.979 |
| chr19:41082647..41082774 | 12.137 | SPTBN4|spectrin, beta, non-erythrocytic 4 | -46279 (+) | SHKBP1|SH3KBP1 binding protein 1 | 39 (+) | |
| chr1:10458961..10459090 | 12.121 | KIF1B|kinesin family member 1B | -188254 (+) | PGD|phosphogluconate dehydrogenase | 104 (+) | 29:0.325 |
| chr1:33502509..33502643 | 12.121 | AK2|adenylate kinase 2 | -76 (-) | ADC|arginine decarboxylase | 44157 (+) | 58:0.778 |
| chr22:39101512..39101696 | 12.068 | JOSD1|Josephin domain containing 1 | -5380 (-) | GTPBP1|GTP binding protein 1 | 261 (+) | -8:0.708 |
| chr14:23938582..23938725 | 12.038 | MYH7|myosin, heavy chain 7, cardiac muscle, beta | -33794 (-) | NGDN|neuroguidin, EIF4E binding protein | 238 (+) | -11:0.987, -53:0.608 |
| chr14:78083185..78083349 | 12.036 | SPTLC2|serine palmitoyltransferase, long chain base subunit 2 | -141 (-) | ALKBH1|alkB, alkylation repair homolog 1 (E. coli) | 91085 (-) | -44:0.546 |
| chr22:37172180..37172348 | 12.024 | IFT27|intraflagellar transport 27 homolog (Chlamydomonas) | -109 (-) | PVALB|parvalbumin | 43255 (-) | -20:0.909, -68:0.476 |
| chr2:74685350..74685506 | 12.014 | INO80B|INO80 complex subunit B | -3224 (+) | WBP1|WW domain binding protein 1 | 199 (+) | 83:0.933 |
| chr7:150725390..150725510 | 11.987 | ATG9B|ATG9 autophagy related 9 homolog B (S. cerevisiae) | -3882 (-) | ABCB8|ATP-binding cassette, sub-family B (MDR/TAP), member 8 | 113 (+) | |
| chr19:19174637..19174762 | 11.977 | ARMC6|armadillo repeat containing 6 | -30039 (+) | SLC25A42|solute carrier family 25, member 42 | 110 (+) | -23:0.260, 56:0.891 |
| chr7:8008243..8008383 | 11.973 | RPA3|replication protein A3, 14kDa | -250076 (-) | GLCCI1|glucocorticoid induced transcript 1 | 120 (+) | 33:0.839, -37:0.556 |
| chr12:112563211..112563321 | 11.949 | NAA25|N(alpha)-acetyltransferase 25, NatB auxiliary subunit | -16668 (-) | TRAFD1|TRAF-type zinc finger domain containing 1 | 56 (+) | 24:0.751, -54:0.675 |
| chr12:58087732..58087916 | 11.937 | B4GALNT1|beta-1,4-N-acetyl-galactosaminyl transferase 1 | -60880 (-) | OS9|osteosarcoma amplified 9, endoplasmic reticulum lectin | 16 (+) | 33:0.524, -77:0.432, -96:0.340 |
| chr5:32313280..32313404 | 11.934 | MTMR12|myotubularin related protein 12 | -209 (-) | ZFR|zinc finger RNA binding protein | 75278 (-) | -72:0.988 |
| chr10:76586193..76586332 | 11.923 | ADK|adenosine kinase | -649675 (+) | KAT6B|K(lysine) acetyltransferase 6B | 92 (+) | |
| chr16:58663824..58663962 | 11.876 | CNOT1|CCR4-NOT transcription complex, subunit 1 | -148 (-) | SLC38A7|solute carrier family 38, member 7 | 54763 (-) | -56:0.426 |
| chr20:44441097..44441208 | 11.849 | DNTTIP1|deoxynucleotidyltransferase, terminal, interacting protein 1 | -20521 (+) | UBE2C|ubiquitin-conjugating enzyme E2C | 129 (+) | 41:0.242, 72:0.290 |
| chrX:3732929..3733089 | 11.758 | PRKX|protein kinase, X-linked | -101291 (-) | NLGN4X|neuroligin 4, X-linked | 2412878 (-) | |
| chr5:31532240..31532381 | 11.731 | DROSHA|drosha, ribonuclease type III | -88 (-) | C5orf22|chromosome 5 open reading frame 22 | 125 (+) | |
| chr11:65382939..65383070 | 11.722 | MAP3K11|mitogen-activated protein kinase kinase kinase 11 | -228 (-) | PCNXL3|pecanex-like 3 (Drosophila) | 702 (+) | |
| chr17:47841565..47841750 | 11.719 | FAM117A|family with sequence similarity 117, member A | -111 (-) | KAT7|K(lysine) acetyltransferase 7 | 24361 (+) | 65:0.923, -88:0.595, 106:0.486 |
| chr19:8455123..8455245 | 11.699 | ANGPTL4|angiopoietin-like 4 | -26077 (+) | RAB11B|RAB11B, member RAS oncogene family | 35 (+) | -25:0.994 |
| chr1:35658790..35658949 | 11.687 | SFPQ|splicing factor proline/glutamine-rich | -124 (-) | ZMYM4|zinc finger, MYM-type 4 | 75693 (+) | 11:0.933 |
| chrX:47420437..47420583 | 11.684 | ZNF41|zinc finger protein 41 | -78166 (-) | ARAF|v-raf murine sarcoma 3611 viral oncogene homolog | 57 (+) | 46:0.494 |
| chr4:775614..775733 | 11.682 | LOC100129917|uncharacterized LOC100129917 | -51 (-) | CPLX1|complexin 1 | 44252 (-) | -16:0.981, 48:0.966 |
| chr12:120638946..120639084 | 11.651 | RPLP0|ribosomal protein, large, P0 | -53 (-) | PXN|paxillin | 48944 (-) | -25:0.459, -54:0.993 |
| chr15:93425638..93425770 | 11.636 | FAM174B|family with sequence similarity 174, member B | -148315 (-) | CHD2|chromodomain helicase DNA binding protein 2 | 17782 (+) | 34:0.227 |
| chr4:52709165..52709271 | 11.633 | CWH43|cell wall biogenesis 43 C-terminal homolog (S. cerevisiae) | -3720946 (+) | DCUN1D4|DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) | 68 (+) | 53:0.385 |
| chr19:55850794..55850957 | 11.609 | TMEM150B|transmembrane protein 150B | -14186 (-) | SUV420H2|suppressor of variegation 4-20 homolog 2 (Drosophila) | 385 (+) | 68:0.652 |
| chr7:77427850..77427996 | 11.599 | TMEM60|transmembrane protein 60 | -176 (-) | PHTF2|putative homeodomain transcription factor 2 | 218 (+) | |
| chr17:7475890..7476026 | 11.556 | SENP3|SUMO1/sentrin/SMT3 specific peptidase 3 | -10529 (+) | EIF4A1|eukaryotic translation initiation factor 4A1 | 134 (+) | |
| chr8:47368577..47368662 | 11.516 | POTEA|POTE ankyrin domain family, member A | -4221014 (+) | KIAA0146|KIAA0146 | 804876 (+) | |
| chr10:5932012..5932132 | 11.500 | ANKRD16|ankyrin repeat domain 16 | -216 (-) | FBXO18|F-box protein, helicase, 18 | 127 (+) | 32:0.689 |
| chr11:105893162..105893305 | 11.475 | KIAA1826|KIAA1826 | -274 (-) | AASDHPPT|aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase | 55132 (+) | |
| chr14:73925284..73925415 | 11.415 | NUMB|numb homolog (Drosophila) | -58 (-) | C14orf169|chromosome 14 open reading frame 169 | 32276 (+) | 3:0.990 |
| chr17:1619644..1619800 | 11.406 | MIR22HG|MIR22 host gene (non-protein coding) | -234 (-) | WDR81|WD repeat domain 81 | 133 (+) | 33:0.332, -38:0.330 |
| chr15:75531972..75532149 | 11.383 | C15orf39|chromosome 15 open reading frame 39 | -37817 (+) | GOLGA6A|golgin A6 family, member A GOLGA6C|golgin A6 family, member C | 18838 (+) | 17:0.650, -23:0.210, 58:0.650, -60:0.650, -101:0.650 |
| chr3:107241683..107241798 | 11.363 | CCDC54|coiled-coil domain containing 54 | -145430 (+) | BBX|bobby sox homolog (Drosophila) | 109 (+) | 24:0.683 |
| chr2:73298466..73298593 | 11.357 | SFXN5|sideroflexin 5 | -93 (-) | SFXN5|sideroflexin 5 | 360 (-) | 0:0.933, 42:0.904, -69:0.909 |
| chr19:19496325..19496445 | 11.329 | -64721 (+) | GATAD2A|GATA zinc finger domain containing 2A | 321 (+) | ||
| chr12:113658900..113659035 | 11.328 | IQCD|IQ motif containing D | -105 (-) | TPCN1|two pore segment channel 1 | 276 (+) | 3:0.978 |
| chrX:58377736..58377828 | 11.313 | ZXDA|zinc finger, X-linked, duplicated A | -440823 (-) | SPIN4|spindlin family, member 4 | 4193417 (-) | |
| chr16:30382259..30382443 | 11.300 | TBC1D10B|TBC1 domain family, member 10B | -827 (-) | MYLPF|myosin light chain, phosphorylatable, fast skeletal muscle | 3759 (+) | 40:0.208, 98:0.972 |
| chr1:143913146..143913271 | 11.289 | FAM72D|family with sequence similarity 72, member D | -78 (-) | PPIAL4A|peptidylprolyl isomerase A (cyclophilin A)-like 4A PPIAL4B|peptidylprolyl isomerase A (cyclophilin A)-like 4B PPIAL4C|peptidylprolyl isomerase A (cyclophilin A)-like 4C PPIAL4G|peptidylprolyl isomerase A (cyclophilin A)-like 4G | 451016 (-) | |
| chr1:153895512..153895680 | 11.261 | GATAD2B|GATA zinc finger domain containing 2B | -145 (-) | DENND4B|DENN/MADD domain containing 4B | 23550 (-) | |
| chr20:40247058..40247168 | 11.255 | CHD6|chromodomain helicase DNA binding protein 6 | -52 (-) | PTPRT|protein tyrosine phosphatase, receptor type, T | 1571397 (-) | |
| chr10:104005062..104005229 | 11.251 | PITX3|paired-like homeodomain 3 | -3916 (-) | GBF1|golgi brefeldin A resistant guanine nucleotide exchange factor 1 | 134 (+) | 45:0.218, -84:0.880, 87:0.748 |
| chr19:8454926..8455097 | 11.230 | ANGPTL4|angiopoietin-like 4 | -25904 (+) | RAB11B|RAB11B, member RAS oncogene family | 208 (+) | |
| chr19:36119753..36119917 | 11.223 | HAUS5|HAUS augmin-like complex, subunit 5 | -16164 (+) | RBM42|RNA binding motif protein 42 | 107 (+) | 19:0.988, 57:0.870 |
| chr9:139743695..139743839 | 11.223 | PHPT1|phosphohistidine phosphatase 1 | -273 (+) | PHPT1|phosphohistidine phosphatase 1 | 100 (+) | -44:0.695 |
| chr8:11659977..11660133 | 11.206 | NEIL2|nei endonuclease VIII-like 2 (E. coli) | -32787 (+) | FDFT1|farnesyl-diphosphate farnesyltransferase 1 | 129 (+) | 67:0.934 |
| chr19:19030302..19030474 | 11.204 | COPE|coatomer protein complex, subunit epsilon | -188 (-) | DDX49|DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 | 92 (+) | -25:0.738, 67:0.366 |
| chr19:50180319..50180451 | 11.200 | BCL2L12|BCL2-like 12 (proline rich) | -11198 (+) | PRMT1|protein arginine methyltransferase 1 | 64 (+) | |
| chr19:58892612..58892741 | 11.196 | ZNF837|zinc finger protein 837 | -260 (-) | RPS5|ribosomal protein S5 | 5976 (+) | |
| chr12:53473198..53473319 | 11.173 | SPRYD3|SPRY domain containing 3 | -71 (-) | IGFBP6|insulin-like growth factor binding protein 6 | 18169 (+) | 12:0.983, -27:0.960 |
| chr18:71959280..71959412 | 11.145 | CYB5A|cytochrome b5 type A (microsomal) | -167 (-) | C18orf63|chromosome 18 open reading frame 63 | 23763 (+) | |
| chr15:64673712..64673847 | 11.134 | KIAA0101|KIAA0101 | -144 (-) | TRIP4|thyroid hormone receptor interactor 4 | 6257 (+) | -59:0.956 |
| chr14:105957345..105957520 | 11.131 | C14orf80|chromosome 14 open reading frame 80 | -913 (+) | C14orf80|chromosome 14 open reading frame 80 | 204 (+) | 0:0.206 |
| chr14:23790521..23790634 | 11.130 | PABPN1|poly(A) binding protein, nuclear 1 | -1181 (+) | PABPN1|poly(A) binding protein, nuclear 1 | 97 (+) | |
| chr16:70323442..70323541 | 11.120 | AARS|alanyl-tRNA synthetase | -80 (-) | DDX19B|DEAD (Asp-Glu-Ala-As) box polypeptide 19B | 9585 (+) | -34:0.959 |
| chr19:2841336..2841459 | 11.051 | ZNF554|zinc finger protein 554 | -21392 (+) | ZNF555|zinc finger protein 555 | 61 (+) | -13:0.497, 18:0.952, -87:0.740 |
| chr16:4852920..4853039 | 11.014 | -81 (-) | GLYR1|glyoxylate reductase 1 homolog (Arabidopsis) | 1924 (-) | 8:0.652, 47:0.626 |
|
| chr1:11322674..11322822 | 10.985 | MTOR|mechanistic target of rapamycin (serine/threonine kinase) | -141 (-) | UBIAD1|UbiA prenyltransferase domain containing 1 | 10568 (+) | -64:0.204, -105:0.360 |
| chr18:14132447..14132599 | 10.982 | ZNF519|zinc finger protein 519 | -66 (-) | ANKRD20A5P|ankyrin repeat domain 20 family, member A5, pseudogene | 46786 (+) | -7:0.848 |
| chr14:68286385..68286520 | 10.960 | ZFYVE26|zinc finger, FYVE domain containing 26 | -3153 (-) | RAD51B|RAD51 homolog B (S. cerevisiae) | 68 (+) | 0:0.979 |
| chr1:24742143..24742307 | 10.943 | C1orf201|chromosome 1 open reading frame 201 | -639 (-) | NIPAL3|NIPA-like domain containing 3 | 78 (+) | |
| chr15:43213053..43213183 | 10.935 | TTBK2|tau tubulin kinase 2 | -136 (-) | UBR1|ubiquitin protein ligase E3 component n-recognin 1 | 185121 (-) | |
| chr1:153940187..153940323 | 10.931 | SLC39A1|solute carrier family 39 (zinc transporter), member 1 | -68 (-) | CREB3L4|cAMP responsive element binding protein 3-like 4 | 119 (+) | |
| chr5:43313807..43313940 | 10.918 | HMGCS1|3-hydroxy-3-methylglutaryl-CoA synthase 1 (soluble) | -277 (-) | CCL28|chemokine (C-C motif) ligand 28 | 83353 (-) | -30:0.260, 30:0.403 |
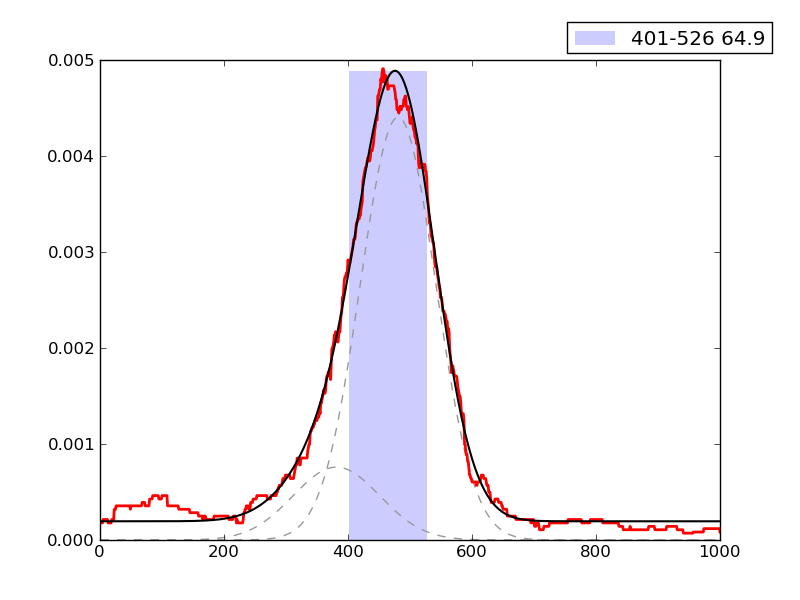
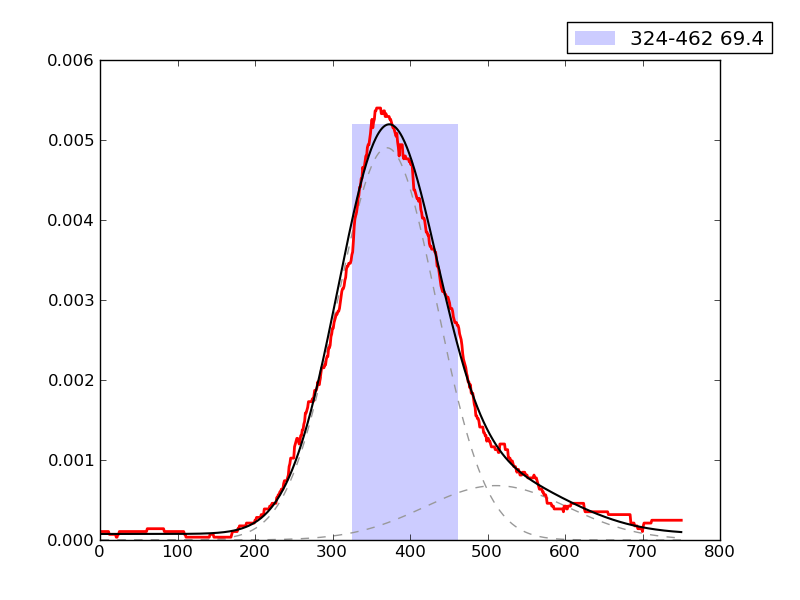
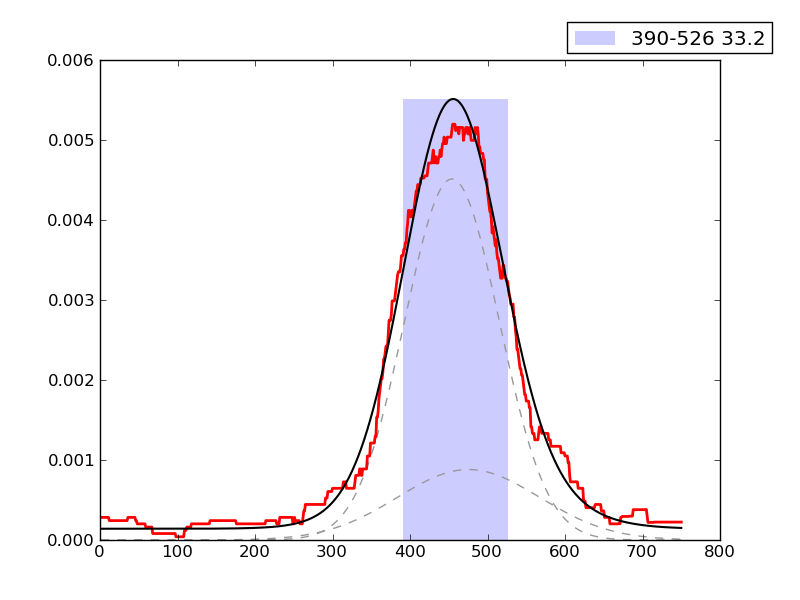
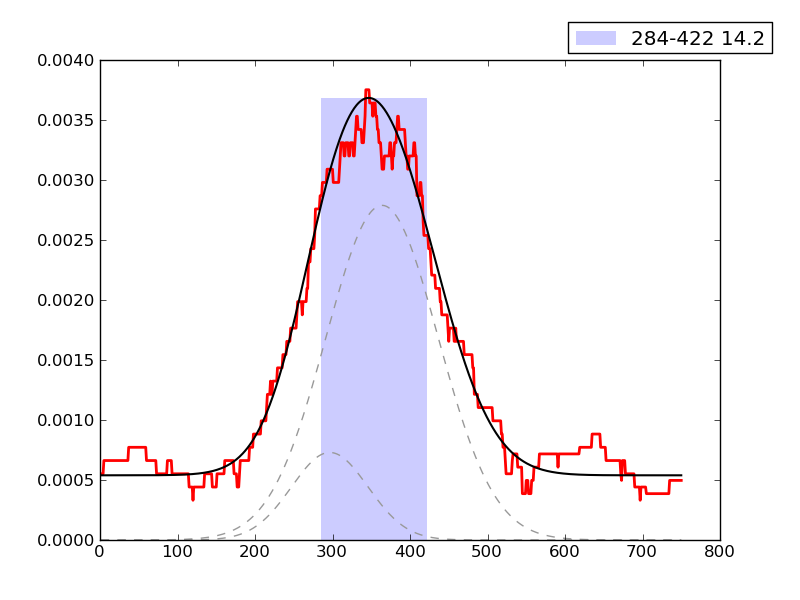
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| denovo_WM_21 | 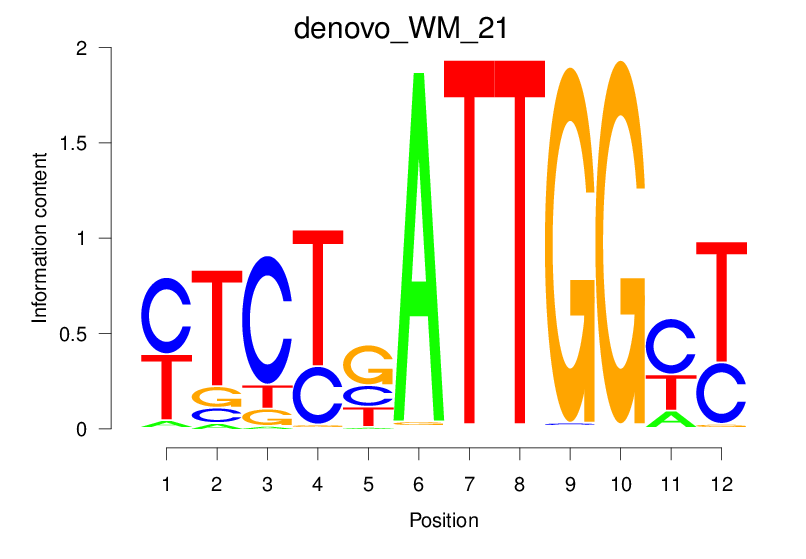 |
24.107 (1591.26) | 24.107 (1591.26) | 0.8348 | 0.257 | 3.4221 | 940/1464 |
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| denovo_WM_21 |  |
24.107 (1591.26) | 0.8348 | 0.257 | 3.4221 | 940/1464 |
| denovo_WM_15 | 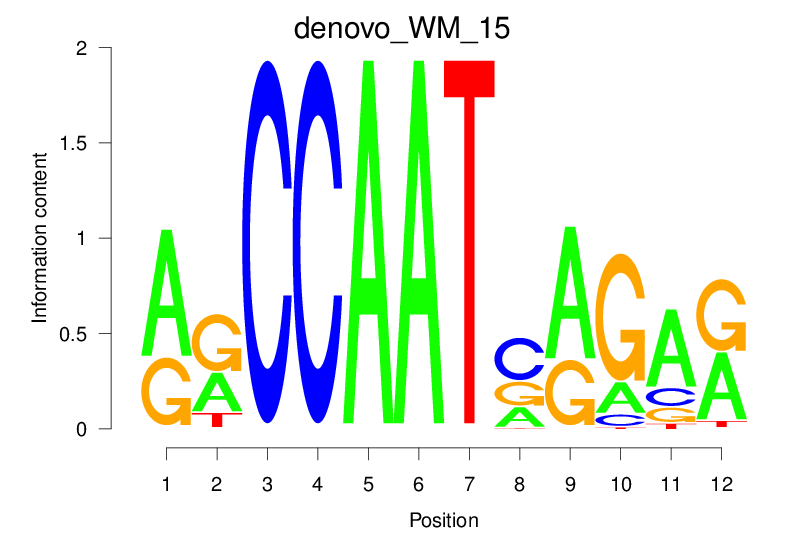 |
23.392 (1576.198) | 0.8263 | 0.2472 | 3.433 | 885/1464 |
| HCMC.NFYB_f1.wm | 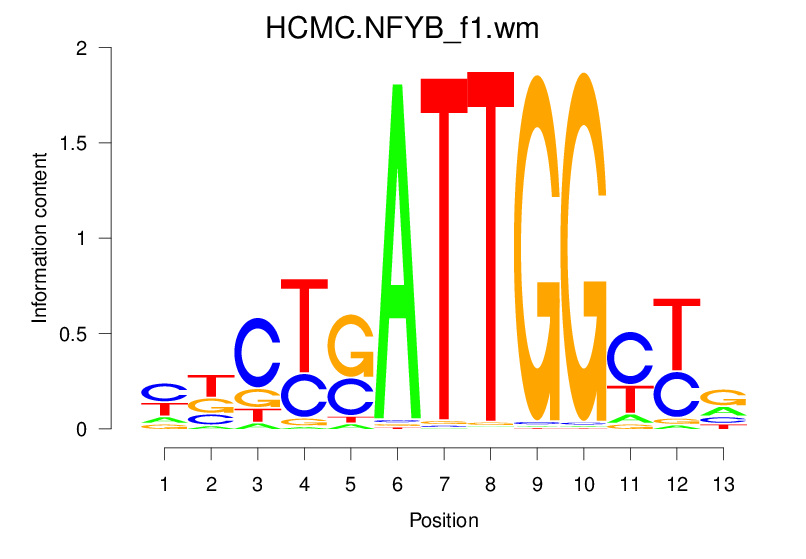 |
19.903 (1495.425) | 0.8668 | 0.281 | 3.2635 | 1158/1464 |
| HCMC.NFYA_f1.wm | 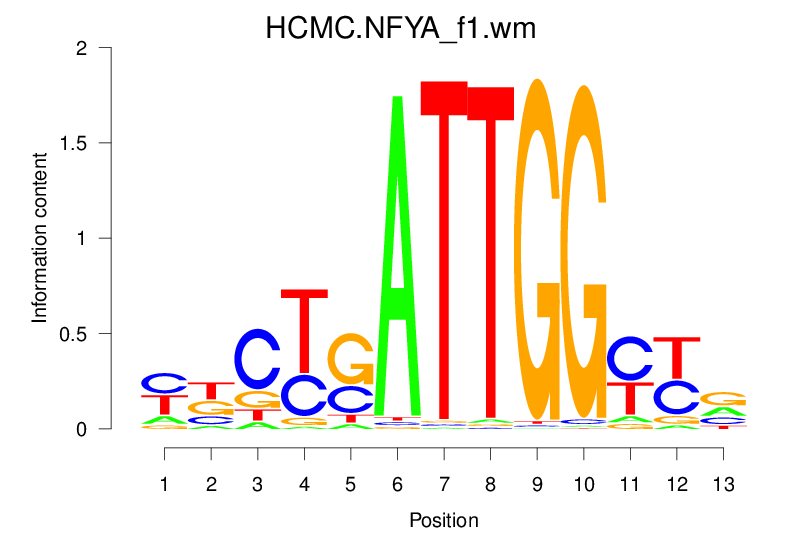 |
18.584 (1461.154) | 0.8770 | 0.2984 | 3.2646 | 1185/1464 |
| denovo_WM_7 | 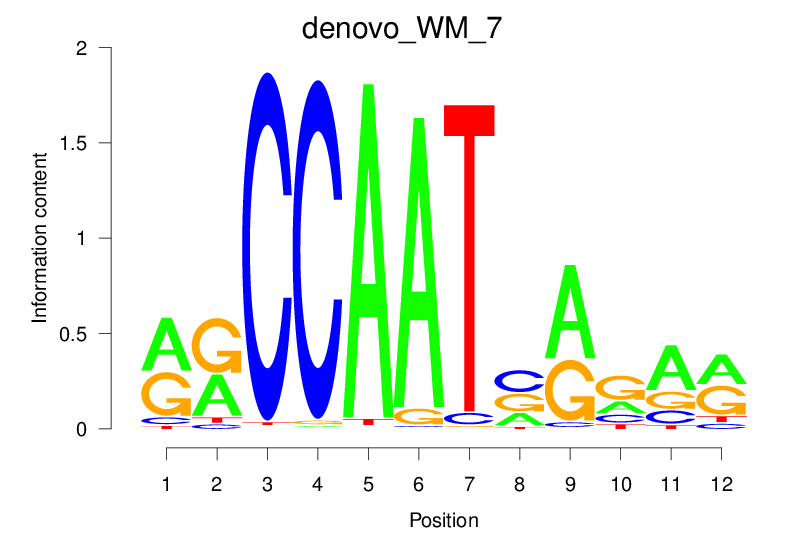 |
18.456 (1457.706) | 0.8785 | 0.3268 | 3.3062 | 1193/1464 |
| denovo_WM_5 | 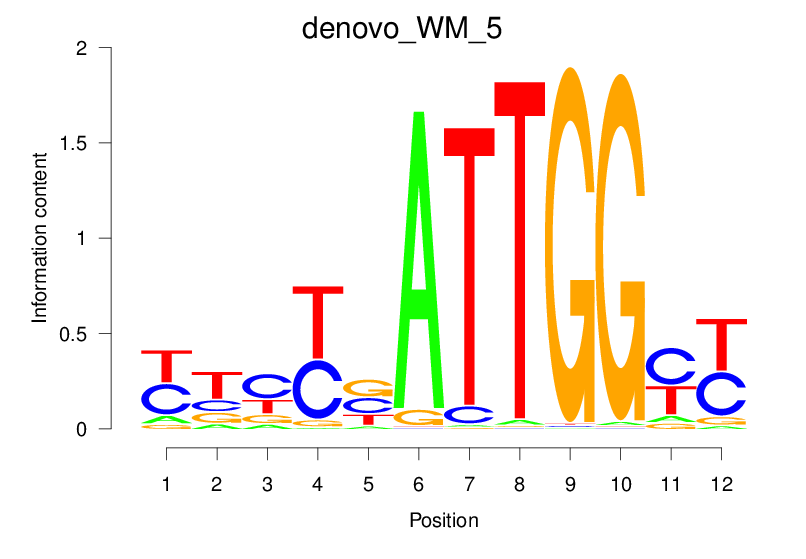 |
17.045 (1417.938) | 0.8831 | 0.3463 | 3.2987 | 1211/1464 |
| JASPAR.NFYA.wm | 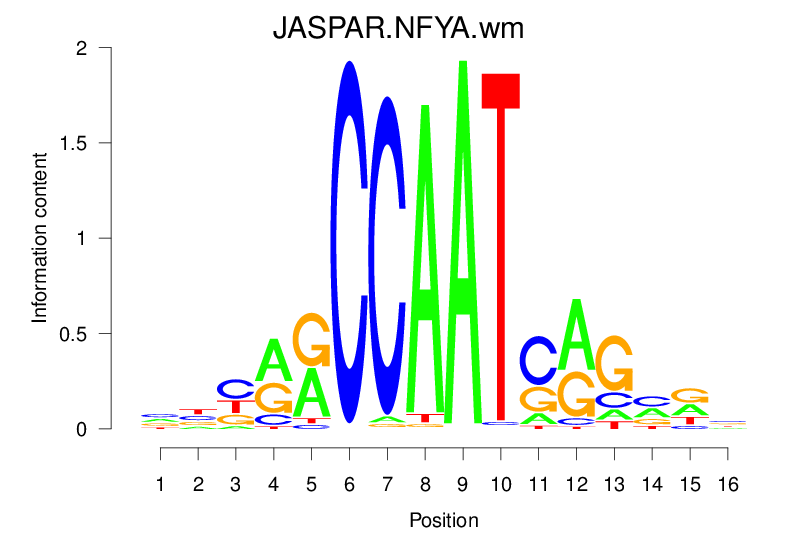 |
16.77 (1409.794) | 0.8742 | 0.2814 | 3.2463 | 1198/1464 |
| denovo_WM_16 | 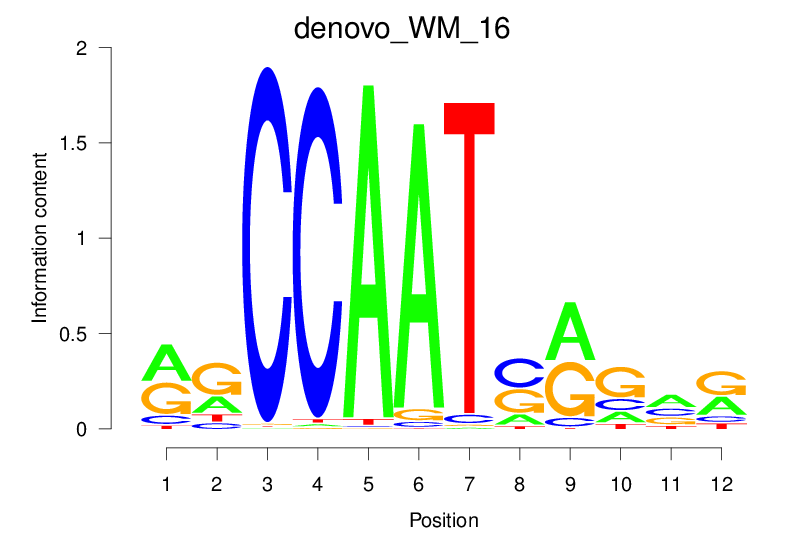 |
15.184 (1360.107) | 0.8901 | 0.3459 | 3.2369 | 1237/1464 |
| denovo_WM_22 | 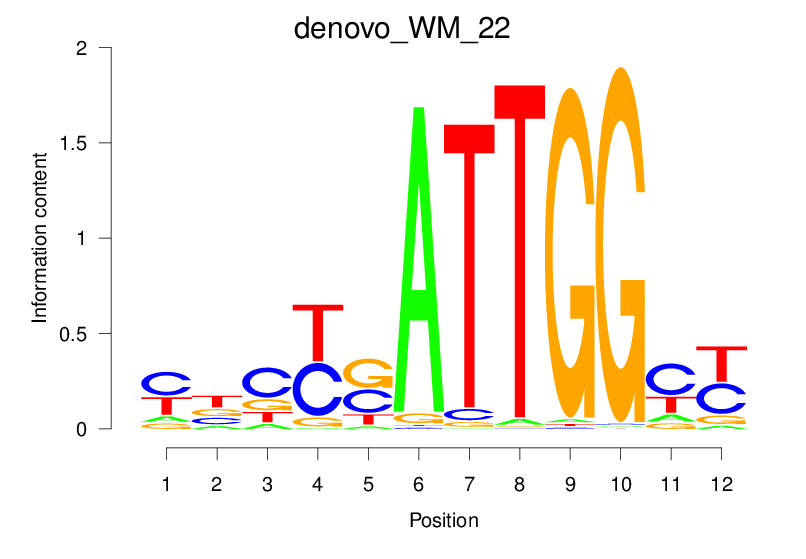 |
14.837 (1348.563) | 0.8901 | 0.3473 | 3.2307 | 1242/1464 |
| denovo_WM_8 | 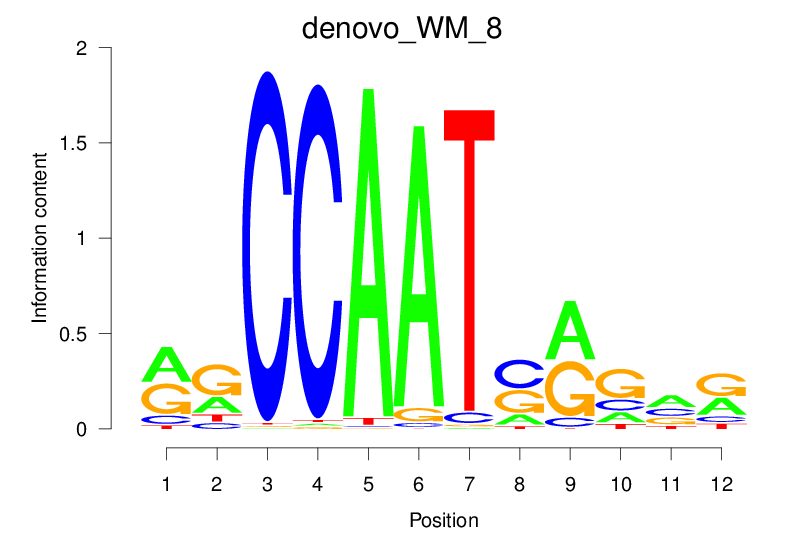 |
14.53 (1338.109) | 0.8903 | 0.3482 | 3.2228 | 1242/1464 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis