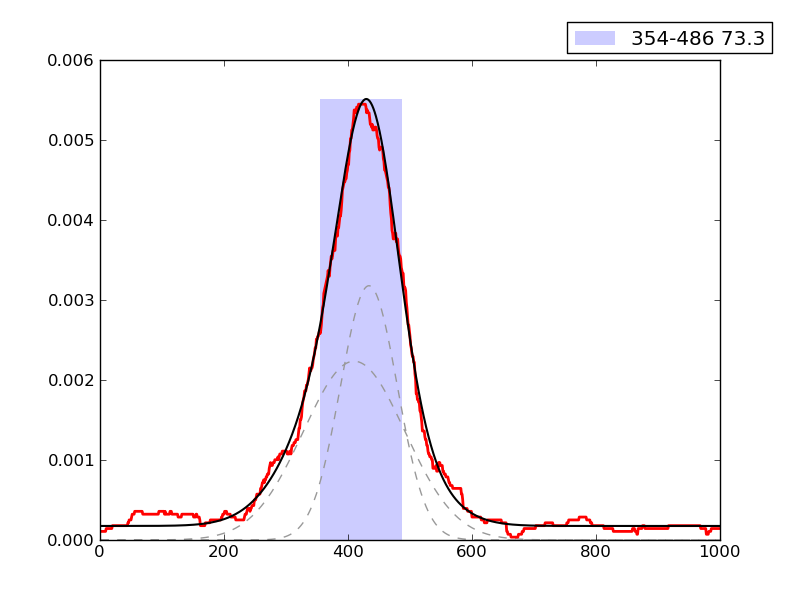
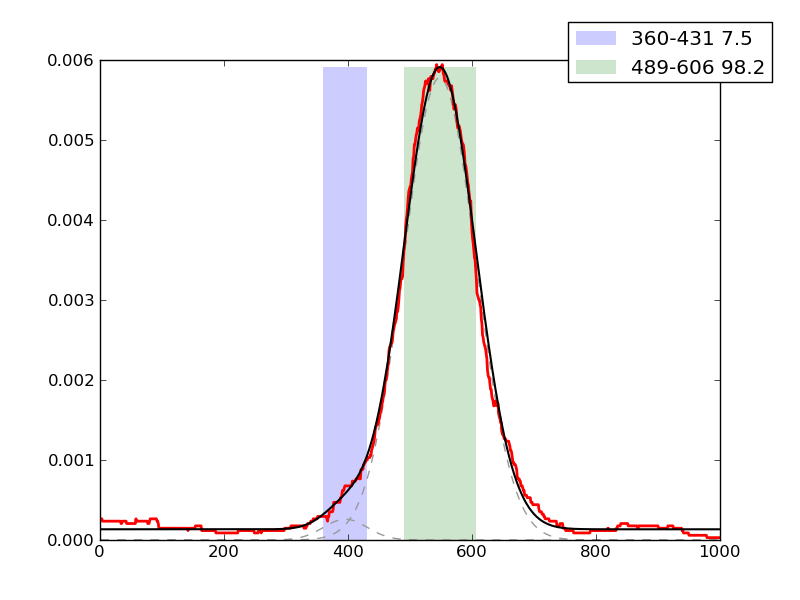
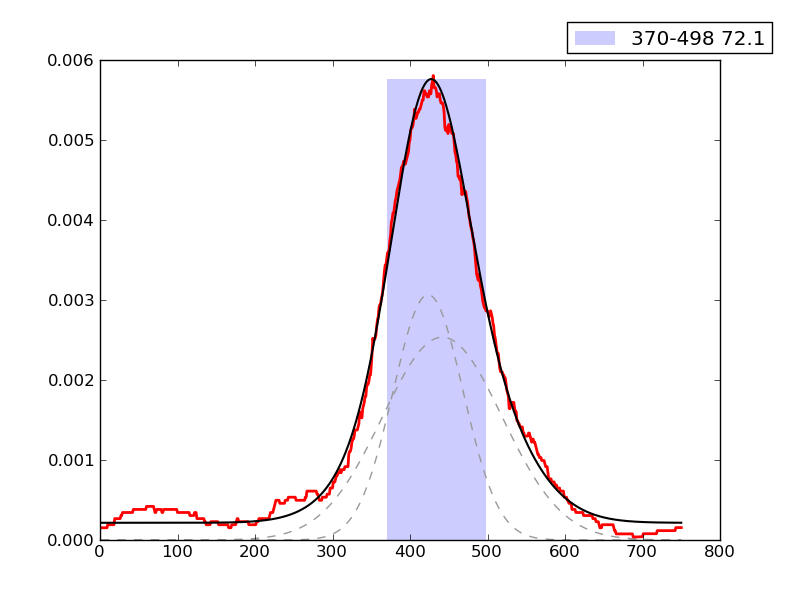
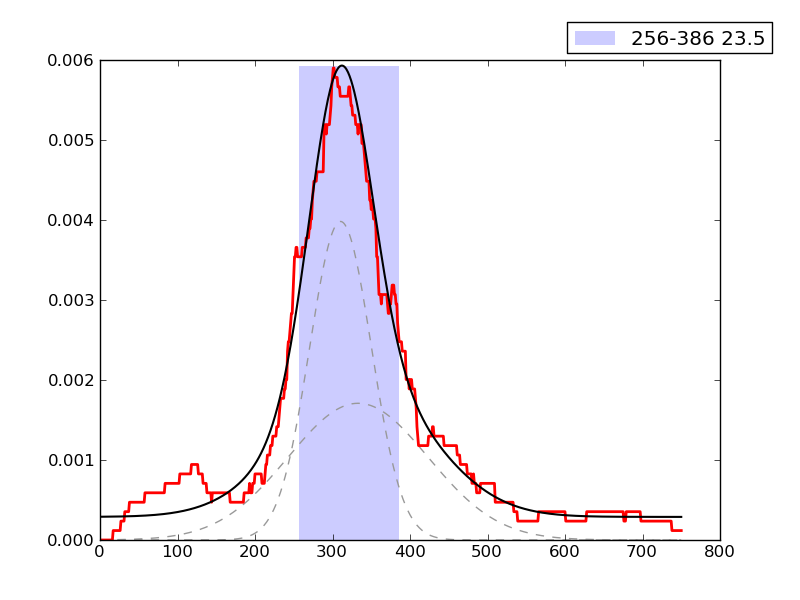
CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.838 | 69th percentile | |
Fraction of mapped reads for the background samples: |
0.890 | 78th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.126 | 13th percentile | |
Error in fit of enrichment distribution: |
0.364 | 13th percentile | |
Peak statistics |
|||
Number of peaks: |
44017 | 89th percentile | |
Fraction of reads mapped to peaks: |
0.056 | 77th percentile | |
Motif enrichment statistics |
|||
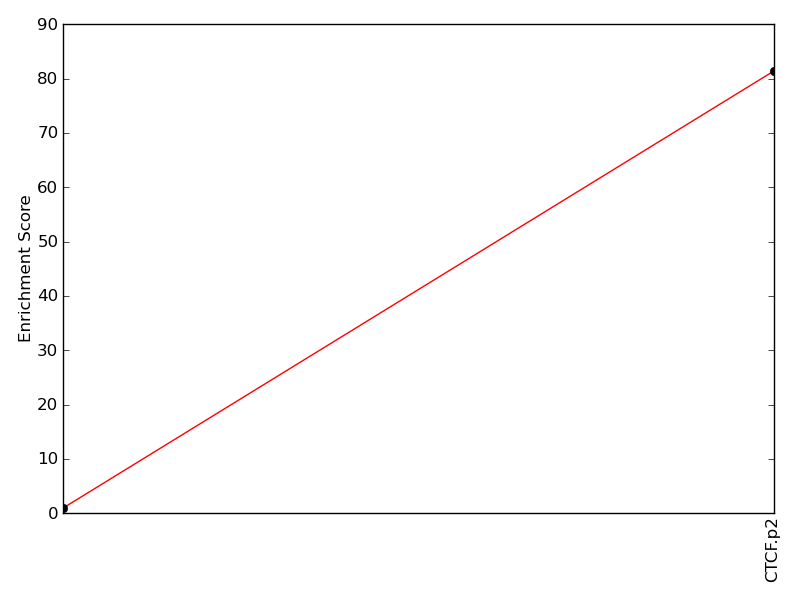
Enrichment of top motif: |
81.40 | 95th percentile | |
Enrichment of complimentary motif set: |
81.40 | 95th percentile | |
Quality Control, Mapping, Fragment Size Estimation:
SMC3 Replicate 1
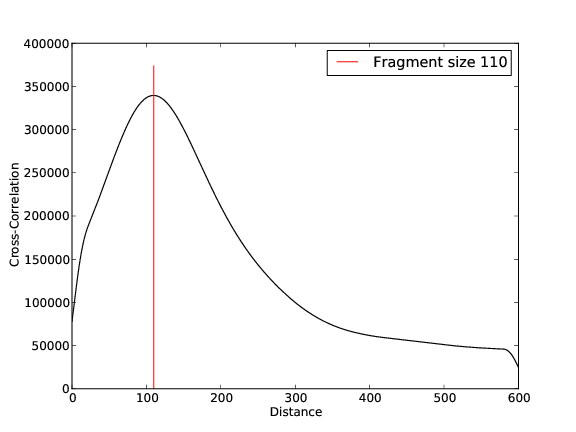
- 31,311,118 input reads (in FASTQ format).
- 28,793,339 reads after removal of low quality reads.
- 28,708,230 reads after removal of adapters and low complexity reads.
- 27,061,910 reads after mapping.
- Report PDF
SMC3 Replicate 2
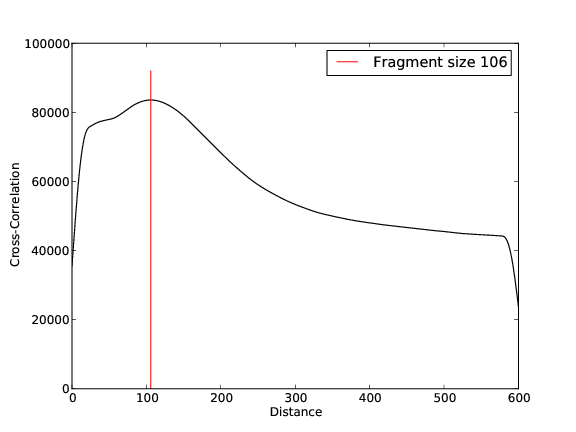
- 42,471,365 input reads (in FASTQ format).
- 36,861,300 reads after removal of low quality reads.
- 36,773,084 reads after removal of adapters and low complexity reads.
- 34,788,972 reads after mapping.
- Report PDF
BG Replicate 1
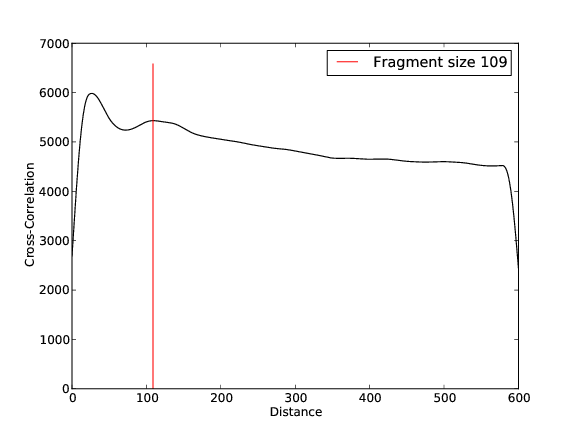
- 6,211,790 input reads (in FASTQ format).
- 6,149,250 reads after removal of low quality reads.
- 6,117,625 reads after removal of adapters and low complexity reads.
- 5,587,889 reads after mapping.
- Report PDF
BG Replicate 2
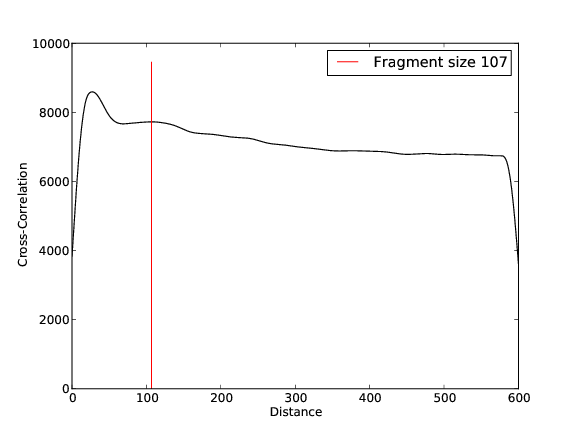
- 5,526,289 input reads (in FASTQ format).
- 5,459,026 reads after removal of low quality reads.
- 5,427,858 reads after removal of adapters and low complexity reads.
- 4,855,998 reads after mapping.
- Report PDF
Peak Calling:
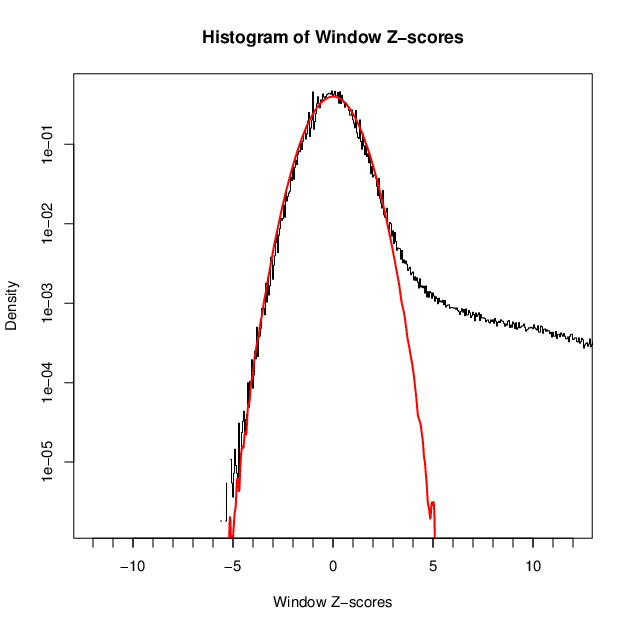
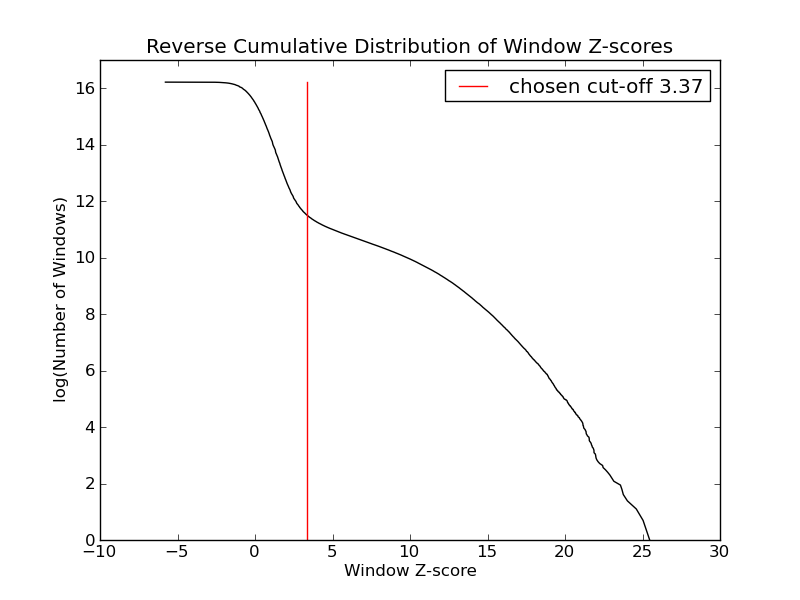
- 98656 windows are significantly enriched at FDR=10% (z-value cut-off: 3.37)
- 98656 windows merged to 47180 regions.
- 44017 significant peaks fitted within 47180 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | CTCF.p2 |
|---|---|---|---|---|---|---|
| chr4:6329117..6329235 | 25.723 | WFS1|Wolfram syndrome 1 (wolframin) | -57550 (+) | PPP2R2C|protein phosphatase 2, regulatory subunit B, gamma | 54408 (-) | 10:1.000 |
| chr7:5603327..5603445 | 24.519 | ACTB|actin, beta | -33177 (-) | FSCN1|fascin homolog 1, actin-bundling protein (Strongylocentrotus purpuratus) | 29052 (+) | -3:0.993 |
| chr3:113007036..113007155 | 24.287 | BOC|Boc homolog (mouse) | -75696 (+) | WDR52|WD repeat domain 52 | 153244 (-) | 4:0.259 |
| chr16:88509554..88509666 | 23.624 | ZNF469|zinc finger protein 469 | -15732 (+) | ZFPM1|zinc finger protein, multitype 1 | 10095 (+) | -13:1.000 |
| chr17:80739793..80739913 | 23.421 | TBCD|tubulin folding cofactor D | -29805 (+) | ZNF750|zinc finger protein 750 | 58056 (-) | -16:1.000 |
| chr19:18107755..18107886 | 23.331 | KCNN1|potassium intermediate/small conductance calcium-activated channel, subfamily N, member 1 | -45710 (+) | ARRDC2|arrestin domain containing 2 | 4130 (+) | 11:1.000 |
| chr1:999732..999912 | 23.300 | LOC100288175|uncharacterized LOC100288175 | -4710 (+) | RNF223|ring finger protein 223 | 9864 (-) | |
| chr15:22436192..22436316 | 23.003 | OR4N4|olfactory receptor, family 4, subfamily N, member 4 | -53809 (+) | 36968 (-) | -31:0.998 |
|
| chr17:6955494..6955633 | 22.953 | SLC16A11|solute carrier family 16, member 11 (monocarboxylic acid transporter 11) | -8322 (-) | CLEC10A|C-type lectin domain family 10, member A | 27988 (-) | -11:0.410 |
| chr17:71251955..71252088 | 22.935 | C17orf80|chromosome 17 open reading frame 80 | -22631 (+) | CPSF4L|cleavage and polyadenylation specific factor 4-like | 5997 (-) | -20:1.000 |
| chr22:50468825..50468943 | 22.836 | IL17REL|interleukin 17 receptor E-like | -17814 (-) | MLC1|megalencephalic leukoencephalopathy with subcortical cysts 1 | 54808 (-) | 8:0.790 |
| chr17:29764425..29764553 | 22.755 | RAB11FIP4|RAB11 family interacting protein 4 (class II) | -45625 (+) | RAB11FIP4|RAB11 family interacting protein 4 (class II) | 50572 (+) | |
| chr8:37773819..37773947 | 22.711 | RAB11FIP1|RAB11 family interacting protein 1 (class I) | -16882 (-) | GOT1L1|glutamic-oxaloacetic transaminase 1-like 1 | 23763 (-) | |
| chr16:85855151..85855300 | 22.628 | COX4I1|cytochrome c oxidase subunit IV isoform 1 | -21890 (+) | IRF8|interferon regulatory factor 8 | 77561 (+) | -50:0.981 |
| chr1:999395..999575 | 22.589 | LOC100288175|uncharacterized LOC100288175 | -4373 (+) | RNF223|ring finger protein 223 | 10201 (-) | |
| chr7:142705138..142705264 | 22.502 | KEL|Kell blood group, metallo-endopeptidase | -45744 (-) | OR9A2|olfactory receptor, family 9, subfamily A, member 2 | 19048 (-) | -9:0.994 |
| chr3:50230164..50230288 | 22.491 | GNAT1|guanine nucleotide binding protein (G protein), alpha transducing activity polypeptide 1 | -1136 (+) | SLC38A3|solute carrier family 38, member 3 | 12480 (+) | -5:0.773 |
| chr17:75491107..75491236 | 22.450 | SEPT9|septin 9 | -19847 (+) | LOC100507351|uncharacterized LOC100507351 | 51849 (+) | 32:0.977 |
| chr11:130731969..130732093 | 22.382 | ADAMTS15|ADAM metallopeptidase with thrombospondin type 1 motif, 15 | -413369 (+) | SNX19|sorting nexin 19 | 54314 (-) | -18:0.955, 36:0.987 |
| chr14:21560769..21560885 | 22.345 | ARHGEF40|Rho guanine nucleotide exchange factor (GEF) 40 | -22393 (+) | ZNF219|zinc finger protein 219 | 5675 (-) | -26:0.691 |
| chr1:23964023..23964150 | 22.314 | ID3|inhibitor of DNA binding 3, dominant negative helix-loop-helix protein | -77802 (-) | RPL11|ribosomal protein L11 | 54194 (+) | 12:1.000 |
| chr16:87812817..87812945 | 22.267 | KLHDC4|kelch domain containing 4 | -13311 (-) | SLC7A5|solute carrier family 7 (amino acid transporter light chain, L system), member 5 | 90194 (-) | -23:0.945 |
| chr12:53278721..53278846 | 22.190 | KRT78|keratin 78 | -36037 (-) | KRT8|keratin 8 | 20033 (-) | -13:0.998 |
| chr22:23301493..23301624 | 22.105 | IGLC1|immunoglobulin lambda constant 1 (Mcg marker) IGLL5|immunoglobulin lambda-like polypeptide 5 | -71475 (+) | GNAZ|guanine nucleotide binding protein (G protein), alpha z polypeptide | 110958 (+) | 20:0.999 |
| chr6:170231878..170232005 | 22.094 | C6orf70|chromosome 6 open reading frame 70 | -80219 (+) | DLL1|delta-like 1 (Drosophila) | 367733 (-) | -22:0.232 |
| chr15:45722556..45722688 | 21.948 | SPATA5L1|spermatogenesis associated 5-like 1 | -28070 (+) | C15orf48|chromosome 15 open reading frame 48 | 175 (+) | -18:0.613, 36:0.996 |
| chr10:45428277..45428406 | 21.947 | TMEM72|transmembrane protein 72 | -21623 (+) | TMEM72-AS1|TMEM72 antisense RNA 1 (non-protein coding) | 26806 (-) | -18:0.993 |
| chr7:75943709..75943831 | 21.933 | HSPB1|heat shock 27kDa protein 1 | -10716 (+) | YWHAG|tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide | 44428 (-) | 36:0.919, -39:0.992 |
| chr1:225662753..225662885 | 21.931 | LBR|lamin B receptor | -46263 (-) | ENAH|enabled homolog (Drosophila) | 177968 (-) | -20:0.998, 32:0.917 |
| chr11:33879524..33879644 | 21.831 | FBXO3|F-box protein 3 | -83592 (-) | LMO2|LIM domain only 2 (rhombotin-like 1) | 11458 (-) | -4:0.999 |
| chr9:139014852..139014976 | 21.801 | C9orf69|chromosome 9 open reading frame 69 | -4185 (-) | LHX3|LIM homeobox 3 | 80074 (-) | 15:0.996 |
| chr4:3445304..3445427 | 21.758 | HGFAC|HGF activator | -1657 (+) | DOK7|docking protein 7 | 19702 (+) | 8:0.467 |
| chr6:110026511..110026649 | 21.730 | FIG4|FIG4 homolog, SAC1 lipid phosphatase domain containing (S. cerevisiae) | -14056 (+) | GPR6|G protein-coupled receptor 6 | 273676 (+) | -37:1.000 |
| chr17:41837860..41837992 | 21.694 | SOST|sclerostin | -1783 (-) | DUSP3|dual specificity phosphatase 3 | 18358 (-) | 35:0.996, -38:0.309 |
| chr15:42103708..42103833 | 21.691 | MAPKBP1|mitogen-activated protein kinase binding protein 1 | -36932 (+) | JMJD7-PLA2G4B|JMJD7-PLA2G4B readthrough JMJD7|jumonji domain containing 7 | 16488 (+) | 19:0.568 |
| chr12:123344917..123345027 | 21.689 | HIP1R|huntingtin interacting protein 1 related | -24919 (+) | 35643 (-) | -3:1.000 |
|
| chr16:57683052..57683175 | 21.669 | GPR56|G protein-coupled receptor 56 | -9796 (+) | GPR97|G protein-coupled receptor 97 | 19094 (+) | -5:0.986, 37:0.326 |
| chr15:78400776..78400902 | 21.655 | SH2D7|SH2 domain containing 7 | -15913 (+) | CIB2|calcium and integrin binding family member 2 | 22923 (-) | 24:1.000 |
| chr15:40567013..40567130 | 21.643 | PAK6|p21 protein (Cdc42/Rac)-activated kinase 6 | -21705 (+) | ANKRD63|ankyrin repeat domain 63 | 7715 (-) | -20:0.990, 21:1.000 |
| chr16:68544195..68544308 | 21.560 | SMPD3|sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | -61770 (-) | ZFP90|zinc finger protein 90 homolog (mouse) | 19782 (+) | 0:0.988 |
| chr22:20960618..20960741 | 21.555 | MED15|mediator complex subunit 15 | -41855 (+) | PI4KA|phosphatidylinositol 4-kinase, catalytic, alpha | 128275 (-) | -6:0.697 |
| chr9:129961617..129961751 | 21.555 | ANGPTL2|angiopoietin-like 2 | -76654 (-) | GARNL3|GTPase activating Rap/RanGAP domain-like 3 | 65123 (+) | |
| chr17:40474901..40475028 | 21.548 | STAT5A|signal transducer and activator of transcription 5A | -34439 (+) | STAT3|signal transducer and activator of transcription 3 (acute-phase response factor) | 65450 (-) | -26:0.999 |
| chr4:173946354..173946484 | 21.528 | GALNTL6|UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 6 | -1211775 (+) | GALNT7|UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 143533 (+) | -6:0.634 |
| chr11:68868992..68869130 | 21.519 | -11535 (+) | MYEOV|myeloma overexpressed (in a subset of t(11;14) positive multiple myelomas) | 192561 (+) | -19:0.965 |
|
| chr7:1032885..1033022 | 21.508 | CYP2W1|cytochrome P450, family 2, subfamily W, polypeptide 1 | -9984 (+) | C7orf50|chromosome 7 open reading frame 50 | 35004 (-) | 22:0.967 |
| chr12:50038458..50038574 | 21.505 | PRPF40B|PRP40 pre-mRNA processing factor 40 homolog B (S. cerevisiae) | -14190 (+) | 62494 (-) | -7:1.000 |
|
| chr10:72309755..72309883 | 21.497 | KIAA1274|KIAA1274 | -71204 (+) | PRF1|perforin 1 (pore forming protein) | 52650 (-) | -16:0.999 |
| chr7:3194636..3194759 | 21.488 | CARD11|caspase recruitment domain family, member 11 | -111235 (-) | SDK1|sidekick cell adhesion molecule 1 | 146302 (+) | 18:0.798 |
| chr12:115100561..115100649 | 21.478 | TBX5|T-box 5 | -254387 (-) | TBX3|T-box 3 | 21535 (-) | -19:0.998 |
| chr1:6306895..6307036 | 21.436 | HES3|hairy and enhancer of split 3 (Drosophila) | -2714 (+) | GPR153|G protein-coupled receptor 153 | 14118 (-) | -9:0.998 |
| chr6:136231046..136231177 | 21.376 | PDE7B|phosphodiesterase 7B | -58260 (+) | FAM54A|family with sequence similarity 54, member A | 340318 (-) | -1:0.852 |
| chr10:102193636..102193780 | 21.354 | LINC00263|long intergenic non-protein coding RNA 263 | -60080 (+) | WNT8B|wingless-type MMTV integration site family, member 8B | 29097 (+) | -27:0.913, 60:0.211 |
| chr8:142441682..142441808 | 21.335 | PTP4A3|protein tyrosine phosphatase type IVA, member 3 | -9684 (+) | FLJ43860|FLJ43860 protein | 75572 (-) | 3:1.000 |
| chr3:46530302..46530435 | 21.323 | LTF|lactotransferrin | -23871 (-) | RTP3|receptor (chemosensory) transporter protein 3 | 9150 (+) | -31:0.627, 32:0.645 |
| chr11:63246422..63246552 | 21.315 | SLC22A9|solute carrier family 22 (organic anion transporter), member 9 | -109093 (+) | HRASLS5|HRAS-like suppressor family, member 5 | 12057 (-) | -20:0.924 |
| chr9:36555943..36556059 | 21.202 | RNF38|ring finger protein 38 | -154826 (-) | MELK|maternal embryonic leucine zipper kinase | 16923 (+) | 14:0.268, 15:0.681 |
| chr2:127285799..127285926 | 21.127 | CNTNAP5|contactin associated protein-like 5 | -2502998 (+) | GYPC|glycophorin C (Gerbich blood group) | 127845 (+) | -6:0.977 |
| chr11:60897350..60897481 | 21.117 | CD5|CD5 molecule | -27436 (+) | VPS37C|vacuolar protein sorting 37 homolog C (S. cerevisiae) | 31515 (-) | 18:0.201, -31:0.799 |
| chr7:73482295..73482401 | 21.096 | ELN|elastin | -39847 (+) | LIMK1|LIM domain kinase 1 | 15778 (+) | -5:0.991 |
| chr7:98062774..98062867 | 21.068 | BAIAP2L1|BAI1-associated protein 2-like 1 | -32387 (-) | NPTX2|neuronal pentraxin II | 183916 (+) | -17:0.744 |
| chr17:61996713..61996847 | 21.063 | GH1|growth hormone 1 | -614 (-) | CD79B|CD79b molecule, immunoglobulin-associated beta | 12886 (-) | -23:0.371 |
| chr22:45131837..45131967 | 21.060 | PRR5-ARHGAP8|PRR5-ARHGAP8 readthrough PRR5|proline rich 5 (renal) | -33649 (+) | ARHGAP8|Rho GTPase activating protein 8 | 16559 (+) | 5:0.999 |
| chr5:133519993..133520099 | 21.041 | SKP1|S-phase kinase-associated protein 1 | -7364 (-) | PPP2CA|protein phosphatase 2, catalytic subunit, alpha isozyme | 41598 (-) | -5:0.993 |
| chr8:28277132..28277277 | 21.038 | ZNF395|zinc finger protein 395 | -33243 (-) | FBXO16|F-box protein 16 ZNF395|zinc finger protein 395 | 70575 (-) | 20:0.931 |
| chr12:53746511..53746645 | 21.015 | SP7|Sp7 transcription factor | -16575 (-) | SP1|Sp1 transcription factor | 27412 (+) | -14:0.988 |
| chr22:41799923..41800060 | 21.014 | TEF|thyrotrophic embryonic factor | -21963 (+) | TOB2|transducer of ERBB2, 2 | 42464 (-) | -10:0.993 |
| chr17:74275674..74275791 | 20.956 | FAM100B|family with sequence similarity 100, member B | -14225 (+) | QRICH2|glutamine rich 2 | 27938 (-) | 16:0.999 |
| chr18:74695380..74695499 | 20.941 | MBP|myelin basic protein | -3409 (-) | MBP|myelin basic protein | 33317 (-) | -4:0.999 |
| chr11:119979019..119979141 | 20.921 | PVRL1|poliovirus receptor-related 1 (herpesvirus entry mediator C) | -379698 (-) | TRIM29|tripartite motif containing 29 | 20466 (-) | 10:0.955 |
| chr17:61502907..61503019 | 20.892 | TANC2|tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 | -231602 (+) | CYB561|cytochrome b-561 | 15227 (-) | -26:0.982 |
| chr2:113484386..113484508 | 20.856 | SLC20A1|solute carrier family 20 (phosphate transporter), member 1 | -80927 (+) | CKAP2L|cytoskeleton associated protein 2-like | 37775 (-) | 32:1.000 |
| chr8:42547760..42547891 | 20.853 | SLC20A2|solute carrier family 20 (phosphate transporter), member 2 | -150764 (-) | CHRNB3|cholinergic receptor, nicotinic, beta 3 | 4800 (+) | -11:0.999 |
| chr17:4316574..4316719 | 20.851 | UBE2G1|ubiquitin-conjugating enzyme E2G 1 | -46738 (-) | SPNS3|spinster homolog 3 (Drosophila) | 20596 (+) | 28:1.000 |
| chr14:70494632..70494767 | 20.850 | SMOC1|SPARC related modular calcium binding 1 | -148570 (+) | SLC8A3|solute carrier family 8 (sodium/calcium exchanger), member 3 | 52193 (-) | 16:0.998 |
| chr1:17563650..17563771 | 20.818 | PADI1|peptidyl arginine deiminase, type I | -32045 (+) | PADI3|peptidyl arginine deiminase, type III | 11889 (+) | 14:1.000 |
| chr17:29852470..29852598 | 20.742 | RAB11FIP4|RAB11 family interacting protein 4 (class II) | -37473 (+) | C17orf79|chromosome 17 open reading frame 79 | 333737 (-) | 15:0.991, 40:0.713 |
| chr15:74731166..74731291 | 20.735 | SEMA7A|semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) | -4916 (-) | UBL7|ubiquitin-like 7 (bone marrow stromal cell-derived) | 22219 (-) | 11:1.000 |
| chr14:105442570..105442690 | 20.718 | AHNAK2|AHNAK nucleoprotein 2 | -5810 (-) | AHNAK2|AHNAK nucleoprotein 2 | 2078 (-) | -8:1.000, 23:0.214 |
| chr9:137335274..137335403 | 20.708 | RXRA|retinoid X receptor, alpha | -116938 (+) | COL5A1|collagen, type V, alpha 1 | 198309 (+) | 8:0.999 |
| chr19:38826240..38826358 | 20.691 | KCNK6|potassium channel, subfamily K, member 6 | -15492 (+) | CATSPERG|cation channel, sperm-associated, gamma | 214 (+) | 1:0.589 |
| chr10:48427008..48427127 | 20.686 | GDF2|growth differentiation factor 2 | -10281 (-) | GDF10|growth differentiation factor 10 | 12012 (-) | -17:0.500 |
| chr2:238421532..238421657 | 20.664 | MLPH|melanophilin | -25698 (+) | PRLH|prolactin releasing hormone | 53622 (+) | -21:0.985 |
| chr15:75063531..75063648 | 20.635 | CYP1A2|cytochrome P450, family 1, subfamily A, polypeptide 2 | -22397 (+) | CSK|c-src tyrosine kinase | 10836 (+) | -4:0.995 |
| chr19:43951307..43951425 | 20.617 | TEX101|testis expressed 101 | -32316 (+) | LYPD3|LY6/PLAUR domain containing 3 | 18410 (-) | -7:0.523 |
| chr19:10691783..10691902 | 20.614 | CDKN2D|cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) | -12204 (-) | AP1M2|adaptor-related protein complex 1, mu 2 subunit | 6106 (-) | |
| chr16:30996380..30996503 | 20.596 | -26904 (+) | HSD3B7|hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 7 | 88 (+) | 0:0.982 |
|
| chr2:10575179..10575293 | 20.579 | HPCAL1|hippocalcin-like 1 | -131411 (+) | ODC1|ornithine decarboxylase 1 | 13124 (-) | 11:0.999 |
| chr15:94867564..94867690 | 20.563 | MCTP2|multiple C2 domains, transmembrane 2 | -26166 (+) | MCTP2|multiple C2 domains, transmembrane 2 | 31682 (+) | 11:1.000 |
| chr20:61640521..61640657 | 20.559 | BHLHE23|basic helix-loop-helix family, member e23 | -2203 (-) | YTHDF1|YTH domain family, member 1 | 206934 (-) | 9:0.999 |
| chr11:2449571..2449690 | 20.502 | TRPM5|transient receptor potential cation channel, subfamily M, member 5 | -5373 (-) | KCNQ1|potassium voltage-gated channel, KQT-like subfamily, member 1 | 16619 (+) | -15:1.000 |
| chr1:205628178..205628299 | 20.480 | ELK4|ELK4, ETS-domain protein (SRF accessory protein 1) | -26239 (-) | SLC45A3|solute carrier family 45, member 3 | 21330 (-) | |
| chr16:67850550..67850682 | 20.468 | TSNAXIP1|translin-associated factor X interacting protein 1 | -9597 (+) | CENPT|centromere protein T | 17149 (-) | -18:1.000, 38:0.475, -69:0.213 |
| chr3:42141381..42141508 | 20.467 | TRAK1|trafficking protein, kinesin binding 1 | -8611 (+) | TRAK1|trafficking protein, kinesin binding 1 | 49304 (+) | 27:0.932 |
| chr1:155908078..155908205 | 20.461 | KIAA0907|KIAA0907 | -3939 (-) | RXFP4|relaxin/insulin-like family peptide receptor 4 | 3348 (+) | -19:0.300 |
| chr2:37641696..37641830 | 20.438 | QPCT|glutaminyl-peptide cyclotransferase | -69950 (+) | CDC42EP3|CDC42 effector protein (Rho GTPase binding) 3 | 257016 (-) | |
| chr14:23590118..23590258 | 20.431 | CEBPE|CCAAT/enhancer binding protein (C/EBP), epsilon | -1369 (-) | SLC7A8|solute carrier family 7 (amino acid transporter light chain, L system), member 8 | 33373 (-) | 32:0.998, -35:1.000 |
| chr1:26471717..26471849 | 20.401 | PDIK1L|PDLIM1 interacting kinase 1 like | -33444 (+) | FAM110D|family with sequence similarity 110, member D | 13757 (+) | 4:0.995 |
| chr17:41771886..41772005 | 20.400 | MEOX1|mesenchyme homeobox 1 | -32670 (-) | SOST|sclerostin | 64198 (-) | |
| chr1:46502847..46502967 | 20.367 | MAST2|microtubule associated serine/threonine kinase 2 | -123645 (+) | PIK3R3|phosphoinositide-3-kinase, regulatory subunit 3 (gamma) | 95571 (-) | -4:1.000 |
| chr17:47472509..47472614 | 20.357 | ZNF652|zinc finger protein 652 | -32981 (-) | PHB|prohibitin | 19654 (-) | 4:1.000 |
| chr8:71446790..71446930 | 20.354 | NCOA2|nuclear receptor coactivator 2 | -130834 (-) | TRAM1|translocation associated membrane protein 1 | 73662 (-) | |
| chr22:20144654..20144767 | 20.349 | LOC388849|uncharacterized LOC388849 | -7280 (-) | RTN4R|reticulon 4 receptor | 111057 (-) | -11:1.000 |
| chr2:238480737..238480864 | 20.341 | PRLH|prolactin releasing hormone | -5584 (+) | RAB17|RAB17, member RAS oncogene family | 18546 (-) | 15:1.000 |
| chr8:143528965..143529089 | 20.325 | TSNARE1|t-SNARE domain containing 1 | -44490 (-) | BAI1|brain-specific angiogenesis inhibitor 1 | 16349 (+) | 3:0.999 |
| chr7:48149870..48149999 | 20.323 | UPP1|uridine phosphorylase 1 | -20979 (+) | ABCA13|ATP-binding cassette, sub-family A (ABC1), member 13 | 61121 (+) | 8:1.000 |
| chr1:9515939..9516083 | 20.321 | -100065 (+) | SLC25A33|solute carrier family 25, member 33 | 83521 (+) | 11:0.767 |
|
| chr1:29208690..29208822 | 20.302 | OPRD1|opioid receptor, delta 1 | -70103 (+) | EPB41|erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) | 4835 (+) | -22:0.986 |
| chr20:32045109..32045238 | 20.294 | SNTA1|syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | -13612 (-) | CBFA2T2|core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | 32663 (+) | -2:0.989 |
| chr16:19897694..19897834 | 20.237 | GPRC5B|G protein-coupled receptor, family C, group 5, member B | -1600 (-) | GPR139|G protein-coupled receptor 139 | 187323 (-) | 2:0.900 |
| chr20:34583044..34583167 | 20.222 | C20orf152|chromosome 20 open reading frame 152 | -26569 (+) | LOC647979|uncharacterized LOC647979 | 55799 (-) | -2:0.938 |
| chr19:16358539..16358665 | 20.212 | AP1M1|adaptor-related protein complex 1, mu 1 subunit | -49852 (+) | KLF2|Kruppel-like factor 2 (lung) | 77039 (+) | -9:0.998 |
| chr7:36331195..36331319 | 20.208 | EEPD1|endonuclease/exonuclease/phosphatase family domain containing 1 | -138202 (+) | KIAA0895|KIAA0895 | 75415 (-) | 9:0.975 |
| chr9:136859639..136859743 | 20.170 | VAV2|vav 2 guanine nucleotide exchange factor | -2245 (-) | BRD3|bromodomain containing 3 | 73502 (-) | 4:0.999 |
| chr20:937211..937314 | 20.158 | ANGPT4|angiopoietin 4 | -40346 (-) | RSPO4|R-spondin 4 | 45655 (-) | 9:0.203 |
| chr4:183859390..183859519 | 20.145 | DCTD|dCMP deaminase | -20820 (-) | WWC2|WW and C2 domain containing 2 | 161024 (+) | -10:0.994 |
| chr6:5143994..5144114 | 20.118 | PPP1R3G|protein phosphatase 1, regulatory subunit 3G | -58335 (+) | LYRM4|LYR motif containing 4 | 116943 (-) | -10:0.999 |
| chr19:42612798..42612907 | 20.115 | ZNF574|zinc finger protein 574 | -32486 (+) | POU2F2|POU class 2 homeobox 2 | 23739 (-) | -28:0.335 |
| chr10:85965474..85965592 | 20.107 | CDHR1|cadherin-related family member 1 | -11080 (+) | LRIT2|leucine-rich repeat, immunoglobulin-like and transmembrane domains 2 | 19780 (-) | |
| chr11:117714822..117714955 | 20.104 | FXYD2|FXYD domain containing ion transport regulator 2 | -16122 (-) | FXYD6|FXYD domain containing ion transport regulator 6 | 32501 (-) | -9:0.992 |
| chr1:201377450..201377570 | 20.094 | LAD1|ladinin 1 | -8769 (-) | TNNI1|troponin I type 1 (skeletal, slow) | 13349 (-) | 7:0.624 |
| chr5:79387663..79387801 | 20.068 | THBS4|thrombospondin 4 | -56568 (+) | SERINC5|serine incorporator 5 | 164153 (-) | -11:0.999 |
| chr16:68834268..68834403 | 20.058 | CDH1|cadherin 1, type 1, E-cadherin (epithelial) | -63024 (+) | TMCO7|transmembrane and coiled-coil domains 7 | 43170 (+) | |
| chr3:45970645..45970769 | 20.051 | CCR9|chemokine (C-C motif) receptor 9 | -42652 (+) | CXCR6|chemokine (C-X-C motif) receptor 6 | 14265 (+) | 24:0.983 |
| chr16:89233964..89234084 | 20.044 | -20078 (+) | CDH15|cadherin 15, type 1, M-cadherin (myotubule) | 4175 (+) | 24:0.920 |
|
| chr2:98313854..98313966 | 20.038 | ACTR1B|ARP1 actin-related protein 1 homolog B, centractin beta (yeast) | -33336 (-) | ZAP70|zeta-chain (TCR) associated protein kinase 70kDa | 16132 (+) | 17:0.996 |
| chr11:36768751..36768877 | 20.031 | RAG2|recombination activating gene 2 | -149012 (-) | LRRC4C|leucine rich repeat containing 4C | 3545780 (-) | -34:0.994, 38:0.999 |
| chr1:205242720..205242842 | 20.030 | TMCC2|transmembrane and coiled-coil domain family 2 | -6147 (+) | NUAK2|NUAK family, SNF1-like kinase, 2 | 48062 (-) | -11:0.995 |
| chr1:21695090..21695217 | 20.023 | ECE1|endothelin converting enzyme 1 | -23173 (-) | NBPF3|neuroblastoma breakpoint family, member 3 | 71529 (+) | 11:0.932 |
| chr20:45901674..45901800 | 20.007 | EYA2|eyes absent homolog 2 (Drosophila) | -378185 (+) | ZMYND8|zinc finger, MYND-type containing 8 | 25832 (-) | -8:0.993 |
| chr17:26821858..26821987 | 19.998 | SLC13A2|solute carrier family 13 (sodium-dependent dicarboxylate transporter), member 2 | -21216 (+) | FOXN1|forkhead box N1 | 29036 (+) | -15:1.000 |
| chr12:5131754..5131868 | 19.992 | KCNA1|potassium voltage-gated channel, shaker-related subfamily, member 1 (episodic ataxia with myokymia) | -112751 (+) | KCNA5|potassium voltage-gated channel, shaker-related subfamily, member 5 | 21308 (+) | -7:0.935 |
| chr3:58518597..58518717 | 19.987 | KCTD6|potassium channel tetramerisation domain containing 6 | -34566 (+) | ACOX2|acyl-CoA oxidase 2, branched chain | 4243 (-) | 3:1.000 |
| chr1:175288875..175289010 | 19.983 | KIAA0040|KIAA0040 | -126912 (-) | TNR|tenascin R (restrictin, janusin) | 423768 (-) | 15:1.000 |
| chr1:27611288..27611401 | 19.982 | WDTC1|WD and tetratricopeptide repeats 1 | -50182 (+) | TMEM222|transmembrane protein 222 | 37307 (+) | 4:0.966 |
| chr20:1294278..1294404 | 19.971 | SDCBP2|syndecan binding protein (syntenin) 2 | -128 (-) | LOC100507495|uncharacterized LOC100507495 | 11710 (+) | -19:0.997 |
| chr3:127371134..127371274 | 19.956 | PODXL2|podocalyxin-like 2 | -23172 (+) | ABTB1|ankyrin repeat and BTB (POZ) domain containing 1 | 20613 (+) | -11:0.887 |
| chr1:20244627..20244753 | 19.950 | -35727 (+) | PLA2G2E|phospholipase A2, group IIE | 5390 (-) | -10:0.983 |
|
| chr20:31240825..31240949 | 19.943 | C20orf203|chromosome 20 open reading frame 203 | -1217 (-) | COMMD7|COMM domain containing 7 | 90366 (-) | 18:0.999 |
| chr17:79075654..79075769 | 19.931 | BAIAP2|BAI1-associated protein 2 | -66706 (+) | AATK|apoptosis-associated tyrosine kinase | 30051 (-) | 26:0.900, -104:0.319 |
| chr16:10603528..10603674 | 19.929 | ATF7IP2|activating transcription factor 7 interacting protein 2 | -80861 (+) | EMP2|epithelial membrane protein 2 | 70889 (-) | 18:0.999 |
| chr19:47940024..47940146 | 19.914 | MEIS3|Meis homeobox 3 | -17322 (-) | SLC8A2|solute carrier family 8 (sodium/calcium exchanger), member 2 | 35062 (-) | -16:0.999 |
| chr8:8130839..8130967 | 19.898 | FLJ10661|family with sequence similarity 86, member A pseudogene | -44761 (+) | SGK223|homolog of rat pragma of Rnd2 | 44985 (-) | -12:0.988 |
| chr11:34196012..34196109 | 19.852 | NAT10|N-acetyltransferase 10 (GCN5-related) | -68810 (+) | ABTB2|ankyrin repeat and BTB (POZ) domain containing 2 | 183495 (-) | -6:0.893 |
| chr1:108668262..108668384 | 19.848 | VAV3|vav 3 guanine nucleotide exchange factor | -160723 (-) | SLC25A24|solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 24 | 67073 (-) | 18:0.993 |
| chr16:67678674..67678802 | 19.831 | CTCF|CCCTC-binding factor (zinc finger protein) | -82344 (+) | RLTPR|RGD motif, leucine rich repeats, tropomodulin domain and proline-rich containing | 288 (+) | 25:0.365 |
| chr11:46911302..46911424 | 19.824 | CKAP5|cytoskeleton associated protein 5 | -43510 (-) | LRP4|low density lipoprotein receptor-related protein 4 | 28699 (-) | -15:0.987 |
| chr14:94406115..94406244 | 19.797 | FAM181A|family with sequence similarity 181, member A | -12471 (+) | ASB2|ankyrin repeat and SOCS box containing 2 | 17587 (-) | -20:0.985, 107:0.393 |
| chr2:30929988..30930119 | 19.787 | LCLAT1|lysocardiolipin acyltransferase 1 | -259905 (+) | CAPN13|calpain 13 | 100240 (-) | 9:0.999 |
| chr17:17763636..17763763 | 19.779 | SREBF1|sterol regulatory element binding transcription factor 1 | -23375 (-) | TOM1L2|target of myb1-like 2 (chicken) | 112042 (-) | 10:0.990 |
| chr3:127689938..127690066 | 19.773 | KBTBD12|kelch repeat and BTB (POZ) domain containing 12 | -48100 (+) | SEC61A1|Sec61 alpha 1 subunit (S. cerevisiae) | 81185 (+) | |
| chr2:48647982..48648115 | 19.769 | FOXN2|forkhead box N2 | -106230 (+) | PPP1R21|protein phosphatase 1, regulatory subunit 21 | 19924 (+) | 7:0.959 |
| chr5:173846668..173846798 | 19.761 | HMP19|HMP19 protein | -373981 (+) | MSX2|msh homeobox 2 | 304879 (+) | -5:0.999 |
| chr17:61556386..61556501 | 19.756 | ACE|angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | -2015 (+) | ACE|angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 5748 (+) | 0:0.999 |
| chr11:117924217..117924355 | 19.754 | IL10RA|interleukin 10 receptor, alpha | -66999 (+) | TMPRSS4|transmembrane protease, serine 4 | 23457 (+) | 11:1.000 |
| chr14:68038113..68038240 | 19.732 | PLEKHH1|pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 | -38203 (+) | PIGH|phosphatidylinositol glycan anchor biosynthesis, class H | 28791 (-) | 8:0.471 |
| chr18:56448916..56449058 | 19.727 | MALT1|mucosa associated lymphoid tissue lymphoma translocation gene 1 | -110142 (+) | ZNF532|zinc finger protein 532 | 81132 (+) | -12:1.000 |
| chr19:10755564..10755672 | 19.719 | SLC44A2|solute carrier family 44, member 2 | -19399 (+) | LOC147727|uncharacterized LOC147727 | 8893 (-) | 13:0.635 |
| chr1:17452381..17452512 | 19.703 | PADI2|peptidyl arginine deiminase, type II | -6539 (-) | PADI1|peptidyl arginine deiminase, type I | 79219 (+) | -17:0.998 |
| chr1:36565328..36565448 | 19.691 | ADPRHL2|ADP-ribosylhydrolase like 2 | -10910 (+) | COL8A2|collagen, type VIII, alpha 2 | 461 (-) | -6:1.000 |
| chr1:9927653..9927778 | 19.674 | CLSTN1|calsyntenin 1 | -43149 (-) | CTNNBIP1|catenin, beta interacting protein 1 | 42594 (-) | 1:0.999 |
| chr11:2326693..2326808 | 19.668 | TSPAN32|tetraspanin 32 | -3364 (+) | CD81|CD81 molecule | 71797 (+) | -8:0.997 |
| chr17:74117614..74117774 | 19.658 | EXOC7|exocyst complex component 7 | -17829 (-) | FOXJ1|forkhead box J1 | 19680 (-) | -10:0.853 |
| chr15:42281850..42281973 | 19.647 | EHD4|EH-domain containing 4 | -17158 (-) | PLA2G4E|phospholipase A2, group IVE | 60989 (-) | -18:0.358 |
| chr11:2440993..2441108 | 19.637 | TSSC4|tumor suppressing subtransferable candidate 4 | -17528 (+) | TRPM5|transient receptor potential cation channel, subfamily M, member 5 | 3207 (-) | 11:0.998 |
| chr16:19097923..19098065 | 19.635 | COQ7|coenzyme Q7 homolog, ubiquinone (yeast) | -18729 (+) | ITPRIPL2|inositol 1,4,5-trisphosphate receptor interacting protein-like 2 | 27254 (+) | -20:0.994 |
| chr1:205225399..205225525 | 19.631 | TMCC2|transmembrane and coiled-coil domain family 2 | -27404 (+) | TMCC2|transmembrane and coiled-coil domain family 2 | 11172 (+) | 16:0.935 |
| chr1:205626793..205626915 | 19.618 | ELK4|ELK4, ETS-domain protein (SRF accessory protein 1) | -24855 (-) | SLC45A3|solute carrier family 45, member 3 | 22714 (-) | 8:0.990 |
| chr10:94194890..94194999 | 19.617 | MARCH5|membrane-associated ring finger (C3HC4) 5 | -143890 (+) | IDE|insulin-degrading enzyme | 62549 (-) | 10:0.930 |
| chr7:128731659..128731810 | 19.593 | TNPO3|transportin 3 | -36529 (-) | SMO|smoothened, frizzled family receptor | 96978 (+) | 1:0.925 |
| chr10:103989638..103989777 | 19.588 | ELOVL3|ELOVL fatty acid elongase 3 | -3565 (+) | PITX3|paired-like homeodomain 3 | 11522 (-) | -12:0.977 |
| chr11:1853790..1853933 | 19.576 | CTSD|cathepsin D | -68626 (-) | SYT8|synaptotagmin VIII | 1856 (+) | -7:1.000 |
| chr21:47240590..47240704 | 19.574 | PCBP3|poly(rC) binding protein 3 | -177018 (+) | PCBP3|poly(rC) binding protein 3 | 29241 (+) | 16:0.996 |
| chr3:52178410..52178533 | 19.567 | DUSP7|dual specificity phosphatase 7 | -88196 (-) | POC1A|POC1 centriolar protein homolog A (Chlamydomonas) | 9959 (-) | 10:0.953 |
| chr11:61103803..61103929 | 19.556 | DAK|dihydroxyacetone kinase 2 homolog (S. cerevisiae) | -1774 (+) | CYBASC3|cytochrome b, ascorbate dependent 3 | 20425 (-) | -3:0.999 |
| chr10:115559479..115559613 | 19.553 | C10orf81|chromosome 10 open reading frame 81 | -34969 (+) | DCLRE1A|DNA cross-link repair 1A | 54370 (-) | -5:0.999 |
| chr8:94834666..94834802 | 19.553 | TMEM67|transmembrane protein 67 | -67620 (+) | PDP1|pyruvate dehyrogenase phosphatase catalytic subunit 1 | 94446 (+) | -9:0.993 |
| chr12:1762105..1762219 | 19.544 | WNT5B|wingless-type MMTV integration site family, member 5B | -23751 (+) | ADIPOR2|adiponectin receptor 2 | 38004 (+) | |
| chr1:41882877..41882997 | 19.543 | SCMH1|sex comb on midleg homolog 1 (Drosophila) | -175101 (-) | EDN2|endothelin 2 | 67382 (-) | 2:1.000 |
| chr9:37913732..37913883 | 19.531 | MCART1|mitochondrial carrier triple repeat 1 | -9441 (-) | SHB|Src homology 2 domain containing adaptor protein B | 155334 (-) | -30:0.231, 86:0.410 |
| chr17:48919337..48919455 | 19.524 | WFIKKN2|WAP, follistatin/kazal, immunoglobulin, kunitz and netrin domain containing 2 | -6792 (+) | TOB1|transducer of ERBB2, 1 | 24300 (-) | 13:0.971 |
| chr6:37659901..37660026 | 19.512 | CCDC167|coiled-coil domain containing 167 | -192267 (-) | MDGA1|MAM domain containing glycosylphosphatidylinositol anchor 1 | 5743 (-) | 4:0.987 |
| chr3:148991016..148991157 | 19.496 | CP|ceruloplasmin (ferroxidase) | -51279 (-) | TM4SF18|transmembrane 4 L six family member 18 | 60316 (-) | -18:0.943, 36:0.501 |
| chr17:20224214..20224339 | 19.490 | SPECC1|sperm antigen with calponin homology and coiled-coil domains 1 | -115842 (+) | CCDC144C|coiled-coil domain containing 144C | 190 (+) | 22:0.999, -25:0.999 |
| chr7:5125182..5125303 | 19.489 | RBAK-LOC389458|RBAK-LOC389458 readthrough RBAK|RB-associated KRAB zinc finger | -39741 (+) | WIPI2|WD repeat domain, phosphoinositide interacting 2 | 104587 (+) | 7:0.999 |
| chr1:206757114..206757249 | 19.488 | RASSF5|Ras association (RalGDS/AF-6) domain family member 5 | -26284 (+) | EIF2D|eukaryotic translation initiation factor 2D | 28574 (-) | 8:0.999 |
| chr16:23193808..23193924 | 19.487 | USP31|ubiquitin specific peptidase 31 | -33317 (-) | SCNN1G|sodium channel, nonvoltage-gated 1, gamma | 186 (+) | |
| chr1:165897461..165897582 | 19.461 | UCK2|uridine-cytidine kinase 2 | -100435 (+) | FAM78B|family with sequence similarity 78, member B | 238433 (-) | -3:0.737 |
| chr15:43517789..43517919 | 19.457 | EPB42|erythrocyte membrane protein band 4.2 | -4576 (-) | TGM5|transglutaminase 5 | 41200 (-) | -10:0.842 |
| chr16:22012428..22012561 | 19.455 | PDZD9|PDZ domain containing 9 | -66 (-) | C16orf52|chromosome 16 open reading frame 52 | 6994 (+) | -11:0.999 |
| chr9:139383199..139383328 | 19.420 | SEC16A|SEC16 homolog A (S. cerevisiae) | -5021 (-) | NOTCH1|notch 1 | 56987 (-) | 0:0.994 |
| chr1:200457928..200458061 | 19.397 | ZNF281|zinc finger protein 281 | -78800 (-) | KIF14|kinesin family member 14 | 131885 (-) | 13:0.890 |
| chr21:35296446..35296582 | 19.385 | ATP5O|ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | -8422 (-) | MRPS6|mitochondrial ribosomal protein S6 SLC5A3|solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 | 149327 (+) | -26:0.946 |
| chr13:27559566..27559703 | 19.375 | GPR12|G protein-coupled receptor 12 | -224781 (-) | USP12|ubiquitin specific peptidase 12 | 186227 (-) | -23:0.885 |
| chr1:248100341..248100477 | 19.368 | OR2T8|olfactory receptor, family 2, subfamily T, member 8 | -16090 (+) | OR2L13|olfactory receptor, family 2, subfamily L, member 13 | 83 (+) | 26:0.970, -69:0.998 |
| chr9:33722223..33722338 | 19.367 | ANXA2P2|annexin A2 pseudogene 2 | -98062 (+) | PRSS3|protease, serine, 3 | 28360 (+) | 23:0.546, -29:0.998 |
| chr9:71199475..71199609 | 19.336 | C9orf71|chromosome 9 open reading frame 71 | -43777 (-) | PIP5K1B|phosphatidylinositol-4-phosphate 5-kinase, type I, beta | 120834 (+) | -30:0.997, 42:0.973 |
| chr14:72219880..72220016 | 19.335 | SIPA1L1|signal-induced proliferation-associated 1 like 1 | -223907 (+) | RGS6|regulator of G-protein signaling 6 | 178868 (+) | 18:0.969 |
| chr2:9954085..9954222 | 19.331 | YWHAQ|tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | -183028 (-) | TAF1B|TATA box binding protein (TBP)-associated factor, RNA polymerase I, B, 63kDa | 29467 (+) | -8:0.983 |
| chr17:72954122..72954238 | 19.327 | OTOP3|otopetrin 3 | -22286 (+) | 14574 (-) | 16:0.934 |
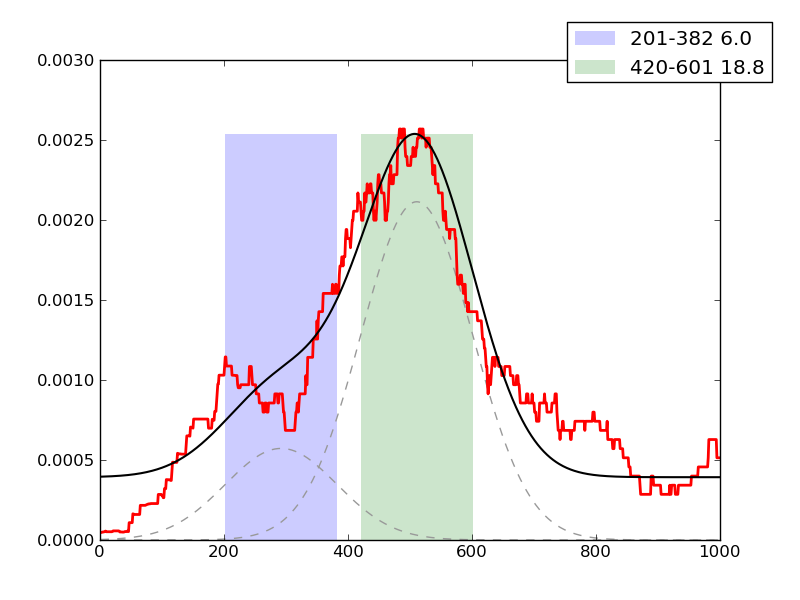
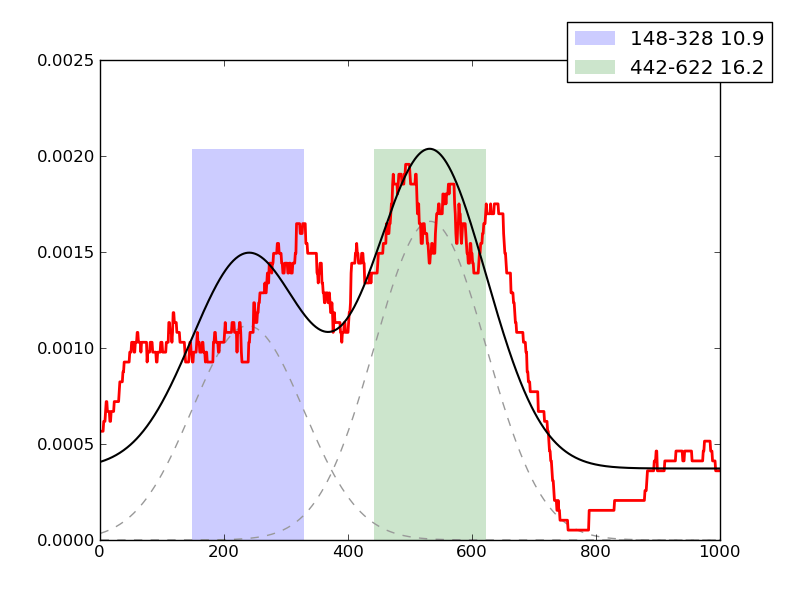
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| CTCF.p2 | 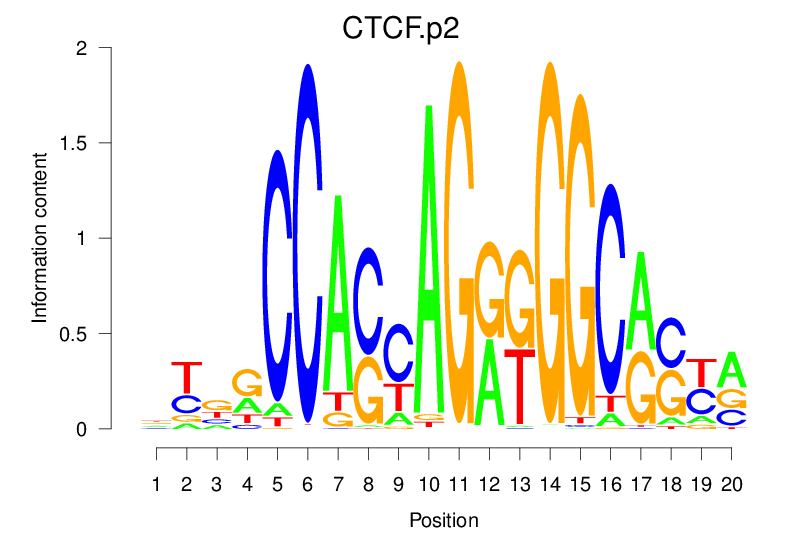 |
81.398 (2199.673) | 81.398 (2199.673) | 0.9491 | 0.4879 | 5.2811 | 25428/44017 |
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| CTCF.p2 |  |
81.398 (2199.673) | 0.9491 | 0.4879 | 5.2811 | 25428/44017 |
| denovo_WM_21 | 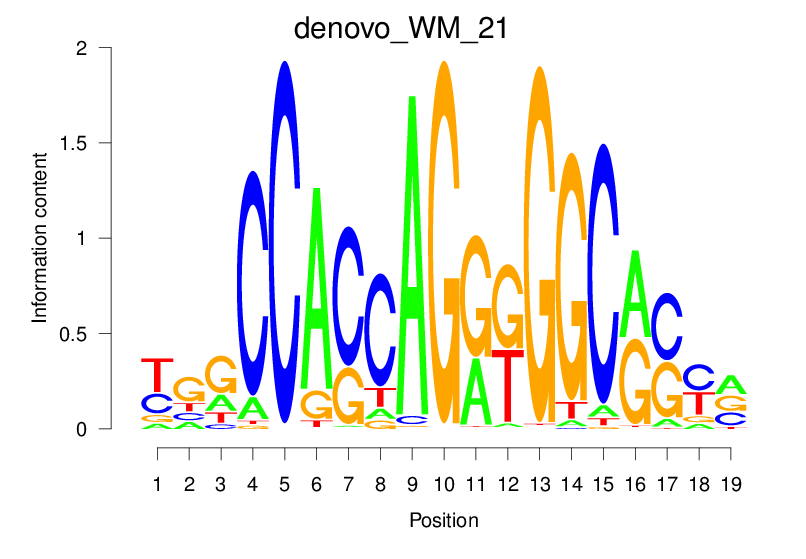 |
58.44 (2034.002) | 0.9418 | 0.4793 | 5.3316 | 24117/44017 |
| denovo_WM_17 | 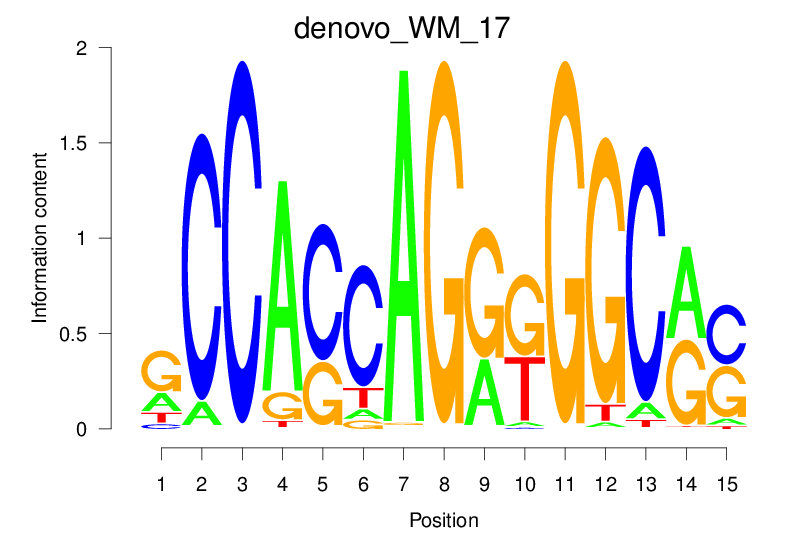 |
52.368 (1979.144) | 0.9236 | 0.4473 | 5.2711 | 22675/44017 |
| JASPAR.CTCF.wm | 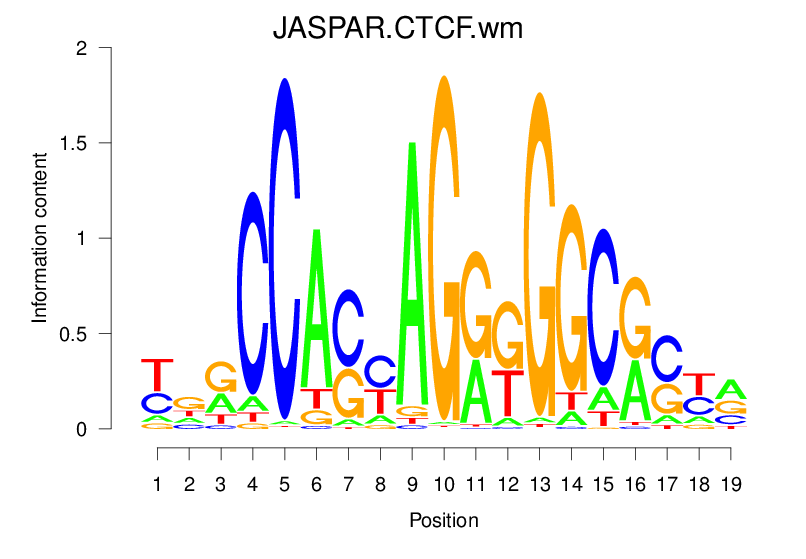 |
36.242 (1795.106) | 0.9505 | 0.4939 | 4.7093 | 30151/44017 |
| HTSELEX.CTCF.C2H2.full.monomeric.wm1 | 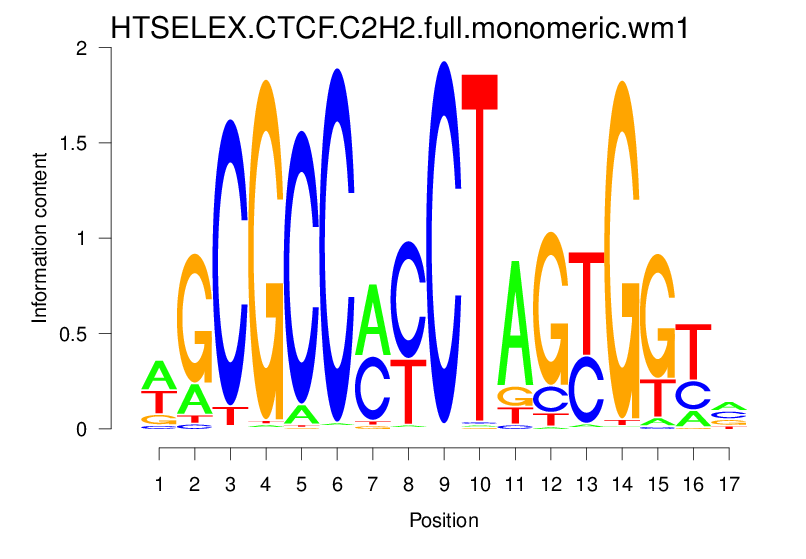 |
22.951 (1566.682) | 0.8098 | 0.3488 | 5.8104 | 13757/44017 |
| denovo_WM_22 | 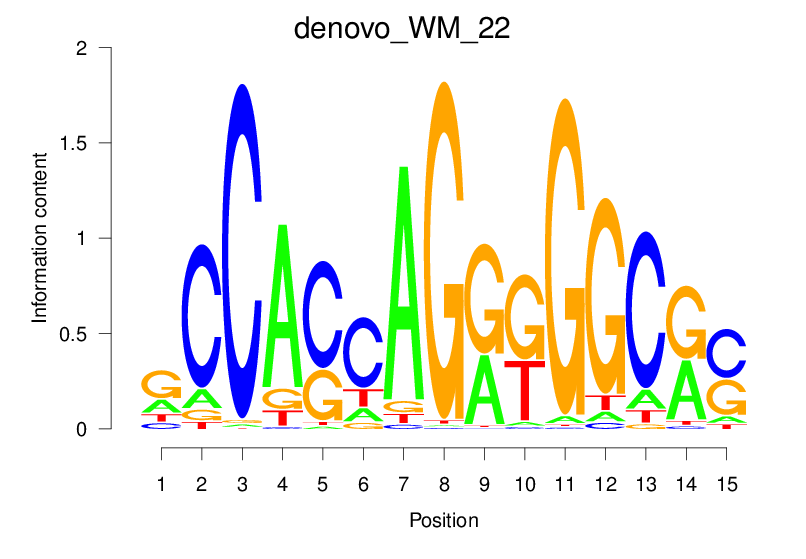 |
22.84 (1564.257) | 0.9325 | 0.4698 | 4.3159 | 30640/44017 |
| denovo_WM_18 | 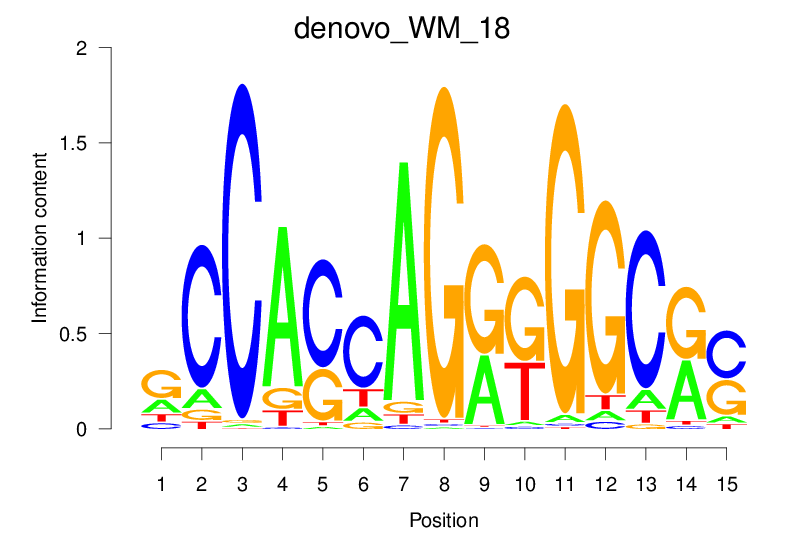 |
22.252 (1551.224) | 0.9328 | 0.4681 | 4.2874 | 30714/44017 |
| HCMC.CTCF_f2.wm | 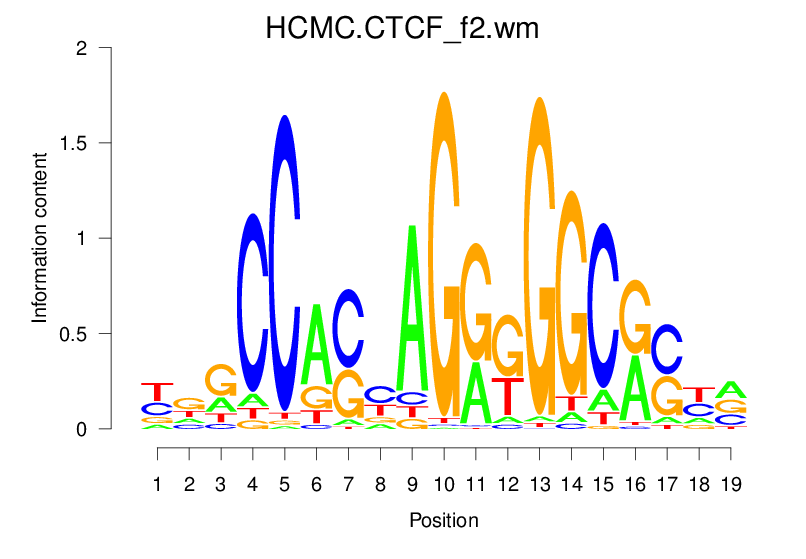 |
18.661 (1463.228) | 0.9403 | 0.4756 | 4.218 | 32154/44017 |
| denovo_WM_5 | 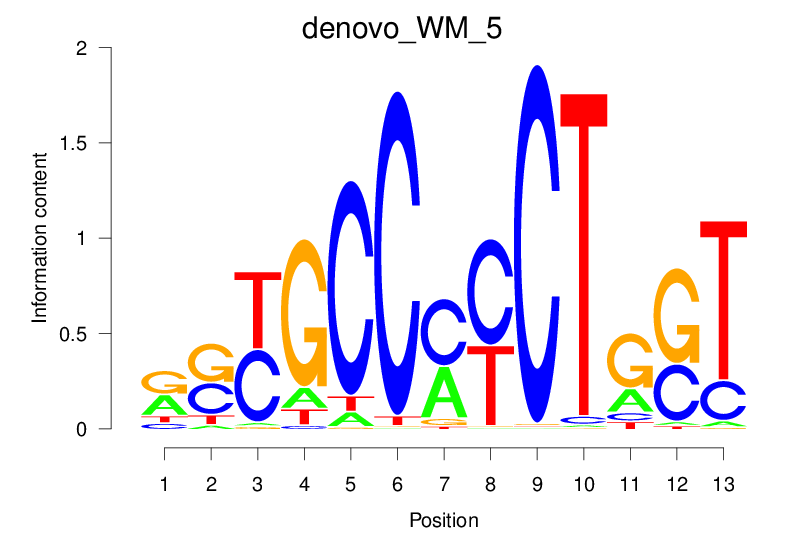 |
10.995 (1198.723) | 0.8173 | 0.4123 | 3.7859 | 27102/44017 |
| denovo_WM_24 | 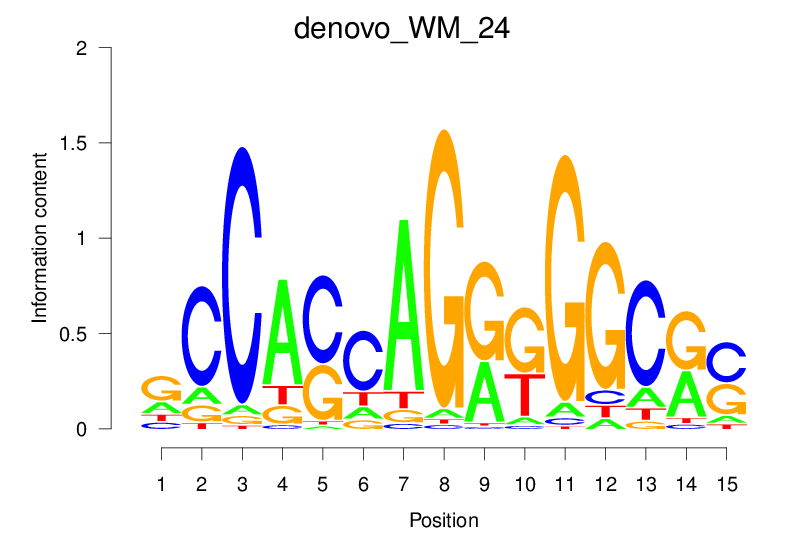 |
10.215 (1161.933) | 0.8914 | 0.414 | 3.258 | 35342/44017 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis