CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.894 | 79th percentile | |
Fraction of mapped reads for the background samples: |
0.890 | 78th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.152 | 26th percentile | |
Error in fit of enrichment distribution: |
0.319 | 5th percentile | |
Peak statistics |
|||
Number of peaks: |
18267 | 59th percentile | |
Fraction of reads mapped to peaks: |
0.024 | 52nd percentile | |
Motif enrichment statistics |
|||
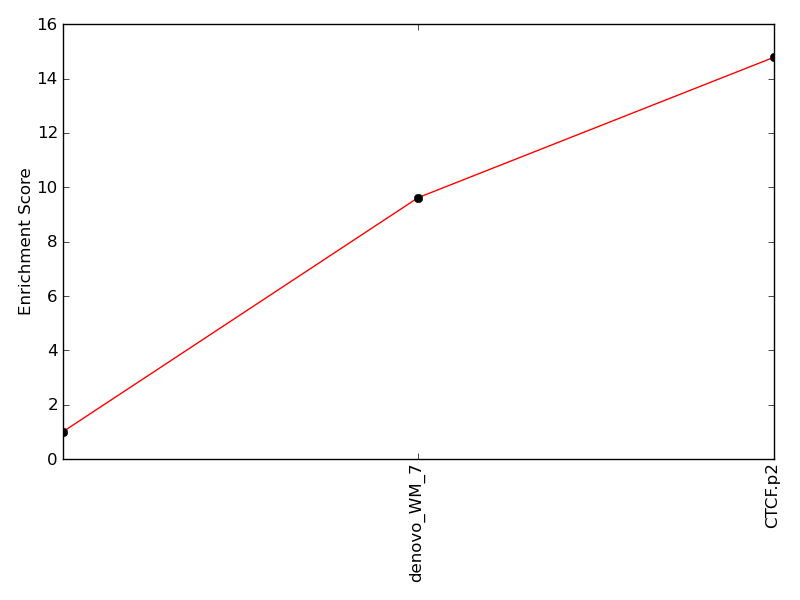
Enrichment of top motif: |
9.62 | 63rd percentile | |
Enrichment of complimentary motif set: |
14.78 | 63rd percentile | |
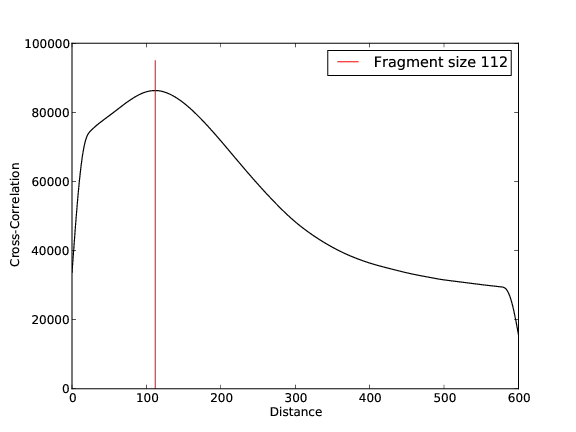
Quality Control, Mapping, Fragment Size Estimation:
ZNF Replicate 1
- 29,142,361 input reads (in FASTQ format).
- 29,019,758 reads after removal of low quality reads.
- 28,868,563 reads after removal of adapters and low complexity reads.
- 26,375,165 reads after mapping.
- Report PDF
ZNF Replicate 2
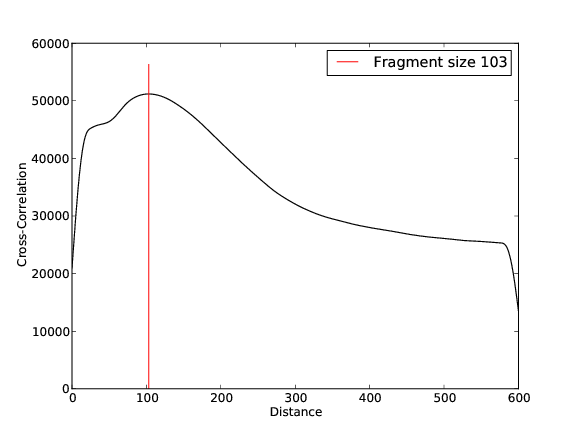
- 29,491,060 input reads (in FASTQ format).
- 29,389,998 reads after removal of low quality reads.
- 29,200,997 reads after removal of adapters and low complexity reads.
- 26,029,452 reads after mapping.
- Report PDF
BG Replicate 1
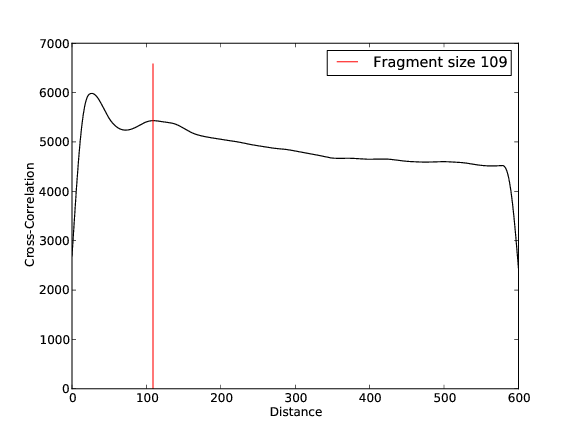
- 6,211,790 input reads (in FASTQ format).
- 6,149,250 reads after removal of low quality reads.
- 6,117,625 reads after removal of adapters and low complexity reads.
- 5,587,889 reads after mapping.
- Report PDF
BG Replicate 2
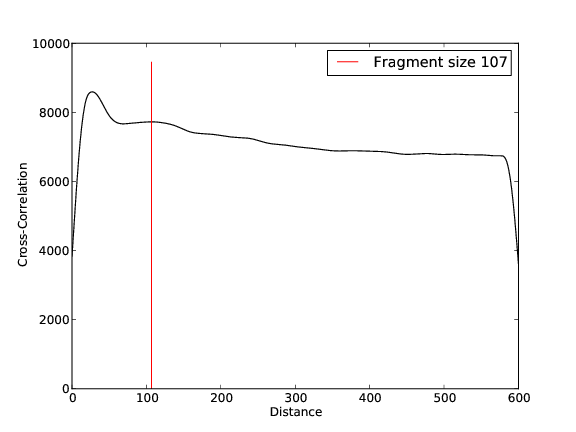
- 5,526,289 input reads (in FASTQ format).
- 5,459,026 reads after removal of low quality reads.
- 5,427,858 reads after removal of adapters and low complexity reads.
- 4,855,998 reads after mapping.
- Report PDF
Peak Calling:
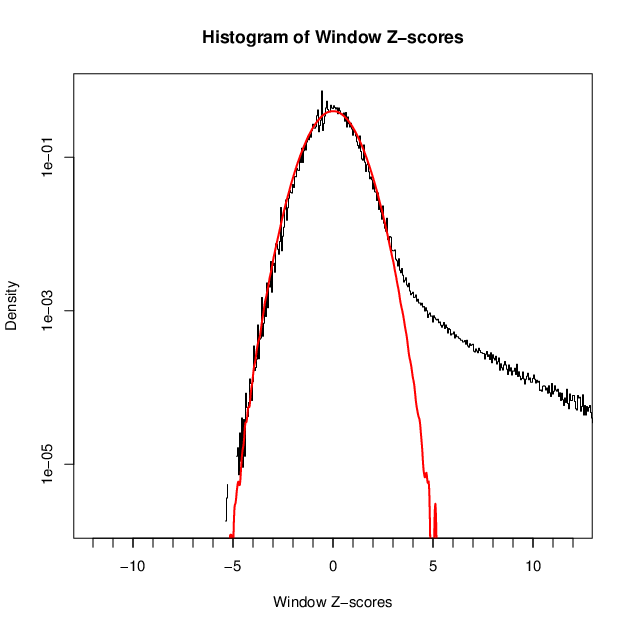
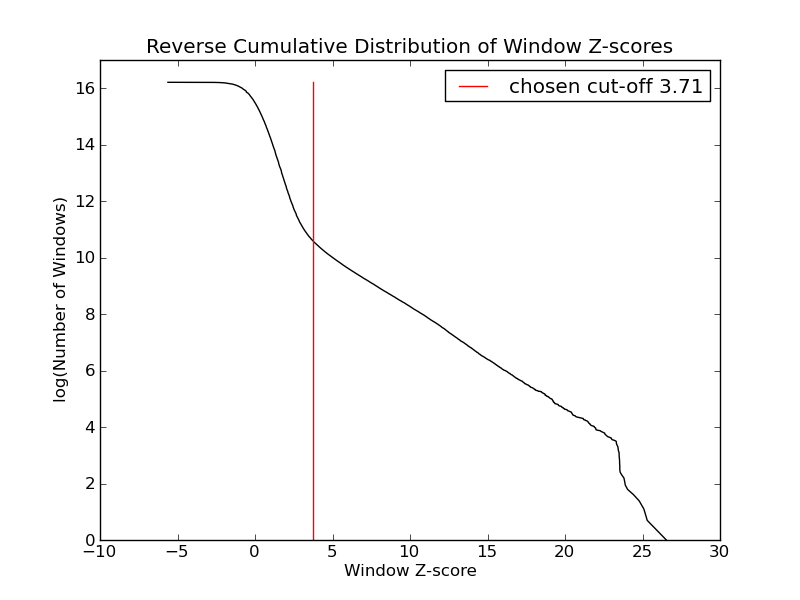
- 40628 windows are significantly enriched at FDR=10% (z-value cut-off: 3.71)
- 40628 windows merged to 20267 regions.
- 18267 significant peaks fitted within 20267 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | denovo_WM_7 | CTCF.p2 |
|---|---|---|---|---|---|---|---|
| chrX:130868823..130868950 | 25.874 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -190888 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 95765 (-) | 105:0.795 |
|
| chrX:130950926..130951053 | 24.853 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -272991 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 13662 (-) | ||
| chrX:52950471..52950599 | 24.718 | FAM156A|family with sequence similarity 156, member A FAM156B|family with sequence similarity 156, member B | -22127 (+) | FAM156A|family with sequence similarity 156, member A FAM156B|family with sequence similarity 156, member B | 35093 (-) | 47:0.454 |
-55:0.927 |
| chrX:52004680..52004804 | 24.448 | MAGED4B|melanoma antigen family D, 4B MAGED4|melanoma antigen family D, 4 | -76765 (+) | XAGE2|X antigen family, member 2 | 107385 (+) | 36:0.982 |
|
| chrX:130845150..130845277 | 24.386 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -167215 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 119438 (-) | ||
| chrX:114960467..114960595 | 24.384 | PLS3|plastin 3 | -132663 (+) | LOC642776|uncharacterized LOC642776 | 124908 (-) | ||
| chrX:115009430..115009550 | 24.121 | PLS3|plastin 3 | -181622 (+) | LOC642776|uncharacterized LOC642776 | 75949 (-) | -56:0.418 |
|
| chrX:114999278..114999405 | 24.085 | PLS3|plastin 3 | -171473 (+) | LOC642776|uncharacterized LOC642776 | 86098 (-) | ||
| chrX:114987320..114987447 | 24.076 | PLS3|plastin 3 | -159515 (+) | LOC642776|uncharacterized LOC642776 | 98056 (-) | ||
| chrX:114972391..114972518 | 24.055 | PLS3|plastin 3 | -144586 (+) | LOC642776|uncharacterized LOC642776 | 112985 (-) | ||
| chrX:114969412..114969539 | 24.045 | PLS3|plastin 3 | -141607 (+) | LOC642776|uncharacterized LOC642776 | 115964 (-) | ||
| chrX:114990307..114990434 | 24.041 | PLS3|plastin 3 | -162502 (+) | LOC642776|uncharacterized LOC642776 | 95069 (-) | ||
| chrX:114996269..114996396 | 24.038 | PLS3|plastin 3 | -168464 (+) | LOC642776|uncharacterized LOC642776 | 89107 (-) | ||
| chrX:114975382..114975509 | 24.033 | PLS3|plastin 3 | -147577 (+) | LOC642776|uncharacterized LOC642776 | 109994 (-) | ||
| chrX:114993276..114993403 | 24.020 | PLS3|plastin 3 | -165471 (+) | LOC642776|uncharacterized LOC642776 | 92100 (-) | ||
| chrX:114978373..114978501 | 24.002 | PLS3|plastin 3 | -150569 (+) | LOC642776|uncharacterized LOC642776 | 107002 (-) | ||
| chr3:183613162..183613292 | 24.000 | PARL|presenilin associated, rhomboid-like | -10519 (-) | ABCC5|ATP-binding cassette, sub-family C (CFTR/MRP), member 5 | 122464 (-) | -21:0.407 |
|
| chrX:114966429..114966556 | 23.987 | PLS3|plastin 3 | -138624 (+) | LOC642776|uncharacterized LOC642776 | 118947 (-) | ||
| chrX:114963448..114963575 | 23.982 | PLS3|plastin 3 | -135643 (+) | LOC642776|uncharacterized LOC642776 | 121928 (-) | ||
| chrX:114984341..114984469 | 23.959 | PLS3|plastin 3 | -156537 (+) | LOC642776|uncharacterized LOC642776 | 101034 (-) | ||
| chrX:114981356..114981483 | 23.958 | PLS3|plastin 3 | -153551 (+) | LOC642776|uncharacterized LOC642776 | 104020 (-) | ||
| chr7:73482265..73482383 | 23.538 | ELN|elastin | -39823 (+) | LIMK1|LIM domain kinase 1 | 15802 (+) | 18:0.991 |
|
| chr11:57434769..57434903 | 23.369 | CLP1|CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae) | -9606 (+) | ZDHHC5|zinc finger, DHHC-type containing 5 | 535 (+) | ||
| chrX:130926451..130926579 | 23.355 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -248517 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 38136 (-) | ||
| chrX:130927266..130927378 | 23.292 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -249324 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 37329 (-) | ||
| chrX:52963970..52964109 | 23.127 | FAM156A|family with sequence similarity 156, member A FAM156B|family with sequence similarity 156, member B | -35631 (+) | FAM156A|family with sequence similarity 156, member A FAM156B|family with sequence similarity 156, member B | 21589 (-) | -49:0.341, -52:0.559 |
52:0.984 |
| chrX:75365485..75365633 | 23.021 | MAGEE2|melanoma antigen family E, 2 | -360531 (-) | CXorf26|chromosome X open reading frame 26 | 27280 (+) | ||
| chr1:46872141..46872256 | 22.994 | FAAH|fatty acid amide hydrolase | -12222 (+) | DMBX1|diencephalon/mesencephalon homeobox 1 | 100474 (+) | 35:0.809, 64:0.268, 65:0.268, 66:0.399 |
-6:0.984 |
| chrX:115002281..115002384 | 22.517 | PLS3|plastin 3 | -174464 (+) | LOC642776|uncharacterized LOC642776 | 83107 (-) | ||
| chr2:238875500..238875620 | 22.502 | RAMP1|receptor (G protein-coupled) activity modifying protein 1 | -107308 (+) | UBE2F|ubiquitin-conjugating enzyme E2F (putative) | 64 (+) | ||
| chr7:129710168..129710321 | 22.420 | ZC3HC1|zinc finger, C3HC-type containing 1 | -19023 (-) | KLHDC10|kelch domain containing 10 | 116 (+) | 2:0.840 |
|
| chrX:75369170..75369289 | 22.304 | MAGEE2|melanoma antigen family E, 2 | -364201 (-) | CXorf26|chromosome X open reading frame 26 | 23610 (+) | ||
| chr17:37617464..37617608 | 22.176 | MED1|mediator complex subunit 1 | -10115 (-) | CDK12|cyclin-dependent kinase 12 | 216 (+) | -11:0.731 |
|
| chrX:130947288..130947416 | 22.109 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -269354 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 17299 (-) | ||
| chr12:122120097..122120217 | 21.818 | MORN3|MORN repeat containing 3 | -12598 (-) | TMEM120B|transmembrane protein 120B | 30587 (+) | 34:0.841, -45:0.454, -61:0.429, -64:0.487 |
-11:0.460 |
| chrX:56804911..56805031 | 21.633 | LOC550643|uncharacterized LOC550643 | -49280 (+) | SPIN3|spindlin family, member 3 | 217000 (-) | 72:0.361 |
|
| chrX:56799512..56799626 | 21.537 | LOC550643|uncharacterized LOC550643 | -43878 (+) | SPIN3|spindlin family, member 3 | 222402 (-) | 73:0.361 |
|
| chrX:130875921..130876042 | 21.382 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -197983 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 88670 (-) | ||
| chrX:115004350..115004480 | 21.289 | PLS3|plastin 3 | -176547 (+) | LOC642776|uncharacterized LOC642776 | 81024 (-) | ||
| chr1:54873343..54873497 | 21.275 | SSBP3|single stranded DNA binding protein 3 | -1336 (-) | ACOT11|acyl-CoA thioesterase 11 | 134541 (+) | 9:0.723, 24:0.732 |
|
| chrX:130875120..130875248 | 21.226 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -197186 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 89467 (-) | ||
| chrX:130912657..130912786 | 20.824 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -234723 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 51930 (-) | ||
| chr10:7453002..7453143 | 20.807 | SFMBT2|Scm-like with four mbt domains 2 | -1822 (-) | SFMBT2|Scm-like with four mbt domains 2 | 362 (-) | -22:0.497, -23:0.490, -37:0.416, -38:0.565 |
|
| chr3:183892324..183892480 | 20.775 | DVL3|dishevelled, dsh homolog 3 (Drosophila) | -19037 (+) | AP2M1|adaptor-related protein complex 2, mu 1 subunit | 205 (+) | -1:0.325, -2:0.261, -19:0.258, -20:0.322 |
|
| chrX:130886538..130886669 | 20.756 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -208605 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 78048 (-) | -104:0.678 |
|
| chr19:10350040..10350218 | 20.747 | S1PR2|sphingosine-1-phosphate receptor 2 | -8123 (-) | MRPL4|mitochondrial ribosomal protein L4 | 12518 (+) | -7:0.739, 25:0.384, 26:0.450 |
|
| chr15:66579892..66580032 | 20.656 | MEGF11|multiple EGF-like-domains 11 | -33885 (-) | DIS3L|DIS3 mitotic control homolog (S. cerevisiae)-like | 5683 (+) | -15:0.541, -30:0.530 |
12:0.272 |
| chr7:18395891..18396036 | 20.605 | HDAC9|histone deacetylase 9 | -65904 (+) | HDAC9|histone deacetylase 9 | 139436 (+) | 2:0.990 |
|
| chr14:23764325..23764437 | 20.581 | HOMEZ|homeobox and leucine zipper encoding | -9056 (-) | BCL2L2-PABPN1|BCL2L2-PABPN1 readthrough BCL2L2|BCL2-like 2 | 11663 (+) | -20:0.811, -63:0.260, -66:0.388 |
8:0.905 |
| chr3:51534090..51534256 | 20.514 | VPRBP|Vpr (HIV-1) binding protein | -167 (-) | RAD54L2|RAD54-like 2 (S. cerevisiae) | 41422 (+) | -11:0.246, -12:0.211, -31:0.209, -32:0.244 |
|
| chrX:130917714..130917842 | 20.413 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -239780 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 46873 (-) | ||
| chr12:106780117..106780236 | 20.412 | POLR3B|polymerase (RNA) III (DNA directed) polypeptide B | -28307 (+) | RFX4|regulatory factor X, 4 (influences HLA class II expression) | 196515 (+) | -48:0.780 |
-1:0.998 |
| chrX:130956443..130956573 | 20.408 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -278510 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 8143 (-) | ||
| chrX:115066551..115066665 | 20.291 | PLS3|plastin 3 | -238740 (+) | LOC642776|uncharacterized LOC642776 | 18831 (-) | ||
| chr1:27721969..27722091 | 20.279 | GPR3|G protein-coupled receptor 3 | -2864 (+) | WASF2|WAS protein family, member 2 | 10154 (-) | -21:0.989 |
41:1.000 |
| chr15:42783332..42783453 | 20.207 | ZFP106|zinc finger protein 106 homolog (mouse) | -68 (-) | SNAP23|synaptosomal-associated protein, 23kDa | 4088 (+) | ||
| chr19:10111036..10111164 | 20.105 | OLFM2|olfactomedin 2 | -63976 (-) | COL5A3|collagen, type V, alpha 3 | 10043 (-) | 35:0.217, 36:0.630, 51:0.660 |
-6:0.837 |
| chrX:130839091..130839200 | 20.084 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -161147 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 125506 (-) | 106:0.224 |
|
| chrX:56794120..56794242 | 20.084 | LOC550643|uncharacterized LOC550643 | -38490 (+) | SPIN3|spindlin family, member 3 | 227790 (-) | 58:0.361 |
|
| chr6:43037810..43037947 | 19.992 | KLC4|kinesin light chain 4 | -9640 (+) | PTK7|PTK7 protein tyrosine kinase 7 | 6199 (+) | -20:0.575, -35:0.468, -36:0.473 |
|
| chrX:130838266..130838401 | 19.935 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -160335 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 126318 (-) | ||
| chr1:160960157..160960273 | 19.910 | ITLN2|intelectin 2 | -35656 (-) | F11R|F11 receptor | 30683 (-) | 10:0.957 |
|
| chr8:1729538..1729663 | 19.811 | CLN8|ceroid-lipofuscinosis, neuronal 8 (epilepsy, progressive with mental retardation) | -17564 (+) | ARHGEF10|Rho guanine nucleotide exchange factor (GEF) 10 | 42539 (+) | -17:0.999 |
|
| chr17:20020068..20020216 | 19.748 | SPECC1|sperm antigen with calponin homology and coiled-coil domains 1 | -29808 (+) | SPECC1|sperm antigen with calponin homology and coiled-coil domains 1 | 39192 (+) | -2:0.860, -17:0.727, -18:0.270 |
36:0.990 |
| chr2:4007033..4007164 | 19.747 | ALLC|allantoicase | -301301 (+) | SOX11|SRY (sex determining region Y)-box 11 | 1825717 (+) | 23:0.297, 24:0.484, 38:0.300, 39:0.452 |
-15:0.251 |
| chrX:130936930..130937063 | 19.547 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -258998 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 27655 (-) | ||
| chr3:127415157..127415287 | 19.540 | ABTB1|ankyrin repeat and BTB (POZ) domain containing 1 | -23405 (+) | MGLL|monoglyceride lipase | 126208 (-) | 25:0.265, 26:0.221, 44:0.231, 45:0.275 |
|
| chrX:115057248..115057376 | 19.520 | PLS3|plastin 3 | -229444 (+) | LOC642776|uncharacterized LOC642776 | 28127 (-) | ||
| chr12:122028707..122028887 | 19.511 | KDM2B|lysine (K)-specific demethylase 2B | -9883 (-) | ORAI1|ORAI calcium release-activated calcium modulator 1 | 35846 (+) | 13:0.896, 28:0.885 |
|
| chr14:102569489..102569638 | 19.511 | HSP90AA1|heat shock protein 90kDa alpha (cytosolic), class A member 1 | -16057 (-) | HSP90AA1|heat shock protein 90kDa alpha (cytosolic), class A member 1 | 36476 (-) | ||
| chr19:2085317..2085477 | 19.502 | MOB3A|MOB kinase activator 3A | -154 (-) | MOB3A|MOB kinase activator 3A | 10868 (-) | ||
| chr19:46320572..46320700 | 19.457 | RSPH6A|radial spoke head 6 homolog A (Chlamydomonas) | -2118 (-) | SYMPK|symplekin | 45808 (-) | 45:0.933, 60:0.946 |
-1:0.941, -92:0.808 |
| chr4:74519485..74519618 | 19.450 | RASSF6|Ras association (RalGDS/AF-6) domain family member 6 | -33332 (-) | IL8|interleukin 8 | 86747 (+) | 4:0.790 |
|
| chrX:130897901..130898031 | 19.392 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -219968 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 66685 (-) | ||
| chr2:30937104..30937254 | 19.327 | LCLAT1|lysocardiolipin acyltransferase 1 | -267031 (+) | CAPN13|calpain 13 | 93114 (-) | 7:0.651, 22:0.650, 37:0.655 |
-42:0.944 |
| chr2:29117252..29117409 | 19.238 | TRMT61B|tRNA methyltransferase 61 homolog B (S. cerevisiae) | -24171 (-) | WDR43|WD repeat domain 43 | 208 (+) | -26:0.736 |
|
| chr5:123006807..123006942 | 19.230 | CSNK1G3|casein kinase 1, gamma 3 | -158992 (+) | ZNF608|zinc finger protein 608 | 1073914 (-) | 24:0.319, 39:0.317 |
-19:0.961 |
| chr13:77599810..77599943 | 19.225 | CLN5|ceroid-lipofuscinosis, neuronal 5 | -33707 (+) | FBXL3|F-box and leucine-rich repeat protein 3 | 1406 (-) | ||
| chr6:151773486..151773618 | 19.213 | C6orf211|chromosome 6 open reading frame 211 | -131 (+) | C6orf211|chromosome 6 open reading frame 211 | 90 (+) | ||
| chrX:130880540..130880660 | 19.207 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -202602 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 84051 (-) | ||
| chrX:130887348..130887462 | 19.139 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -209407 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 77246 (-) | 102:0.847 |
|
| chr12:56967264..56967401 | 19.035 | RBMS2|RNA binding motif, single stranded interacting protein 2 | -51635 (+) | BAZ2A|bromodomain adjacent to zinc finger domain, 2A | 39584 (-) | -48:0.852, -63:0.433, -64:0.518 |
14:0.993 |
| chr16:74700967..74701147 | 18.994 | RFWD3|ring finger and WD repeat domain 3 | -280 (-) | MLKL|mixed lineage kinase domain-like | 33680 (-) | -3:0.315, -4:0.315, -5:0.315, -27:0.690 |
|
| chr6:160141782..160141935 | 18.953 | SOD2|superoxide dismutase 2, mitochondrial | -27496 (-) | WTAP|Wilms tumor 1 associated protein | 6314 (+) | 28:0.234, 43:0.233 |
-12:0.977 |
| chr12:54585434..54585550 | 18.909 | SMUG1|single-strand-selective monofunctional uracil-DNA glycosylase 1 | -2806 (-) | CBX5|chromobox homolog 5 | 67832 (-) | ||
| chr12:57881602..57881761 | 18.877 | ARHGAP9|Rho GTPase activating protein 9 | -8051 (-) | MARS|methionyl-tRNA synthetase | 111 (+) | ||
| chrX:130862710..130862845 | 18.876 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -184779 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 101874 (-) | ||
| chr19:10491291..10491443 | 18.820 | TYK2|tyrosine kinase 2 | -88 (-) | CDC37|cell division cycle 37 homolog (S. cerevisiae) | 22802 (-) | -19:0.591, -20:0.216 |
|
| chr1:27481718..27481898 | 18.815 | SLC9A1|solute carrier family 9 (sodium/hydrogen exchanger), member 1 | -347 (-) | WDTC1|WD and tetratricopeptide repeats 1 | 79226 (+) | ||
| chr19:3987905..3988050 | 18.806 | EEF2|eukaryotic translation elongation factor 2 | -2515 (-) | PIAS4|protein inhibitor of activated STAT, 4 | 19752 (+) | 31:0.807, 46:0.807 |
-14:0.332 |
| chr7:74071915..74072064 | 18.787 | GTF2IRD1|GTF2I repeat domain containing 1 | -203554 (+) | GTF2I|general transcription factor IIi | 32 (+) | -3:0.713, 11:0.662, 27:0.806 |
|
| chr12:122028177..122028293 | 18.717 | KDM2B|lysine (K)-specific demethylase 2B | -9321 (-) | ORAI1|ORAI calcium release-activated calcium modulator 1 | 36408 (+) | -64:0.345, -65:0.345, -66:0.276 |
|
| chr14:54955753..54955913 | 18.699 | GMFB|glia maturation factor, beta | -78 (-) | CGRRF1|cell growth regulator with ring finger domain 1 | 20778 (+) | 11:0.562, 12:0.305, 26:0.262, 27:0.519 |
|
| chrX:130902366..130902496 | 18.662 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -224433 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 62220 (-) | ||
| chrX:123094450..123094630 | 18.606 | STAG2|stromal antigen 2 | -16 (+) | STAG2|stromal antigen 2 | 1033 (+) | -45:0.273, -48:0.485, -63:0.397, -64:0.205, -78:0.297, -79:0.501 |
|
| chr8:76314396..76314530 | 18.591 | CRISPLD1|cysteine-rich secretory protein LCCL domain containing 1 | -417554 (+) | HNF4G|hepatocyte nuclear factor 4, gamma | 5725 (+) | -27:0.709 |
|
| chr11:68041240..68041384 | 18.588 | C11orf24|chromosome 11 open reading frame 24 | -1854 (-) | LRP5|low density lipoprotein receptor-related protein 5 | 38777 (+) | -66:0.859, 83:0.298, 84:0.298, 85:0.298 |
|
| chr12:76328412..76328573 | 18.505 | KRR1|KRR1, small subunit (SSU) processome component, homolog (yeast) | -423073 (-) | PHLDA1|pleckstrin homology-like domain, family A, member 1 | 96055 (-) | ||
| chr19:47343892..47344017 | 18.463 | SLC1A5|solute carrier family 1 (neutral amino acid transporter), member 5 | -52232 (-) | AP2S1|adaptor-related protein complex 2, sigma 1 subunit | 10177 (-) | -13:0.821 |
|
| chr10:79789307..79789447 | 18.450 | POLR3A|polymerase (RNA) III (DNA directed) polypeptide A, 155kDa | -63 (-) | RPS24|ribosomal protein S24 | 4179 (+) | 1:0.234, 2:0.234, 3:0.234, 4:0.234 |
32:0.287 |
| chr11:45315279..45315406 | 18.389 | SYT13|synaptotagmin XIII | -7592 (-) | CHST1|carbohydrate (keratan sulfate Gal-6) sulfotransferase 1 | 371842 (-) | 52:0.486, 67:0.487 |
-5:0.390 |
| chr1:161195620..161195780 | 18.384 | APOA2|apolipoprotein A-II | -2302 (-) | TOMM40L|translocase of outer mitochondrial membrane 40 homolog (yeast)-like | 157 (+) | ||
| chr10:88699233..88699349 | 18.378 | BMPR1A|bone morphogenetic protein receptor, type IA | -182804 (+) | MMRN2|multimerin 2 | 18080 (-) | -47:0.433, -48:0.289, -63:0.320, -64:0.465 |
64:0.742 |
| chr5:10250042..10250213 | 18.362 | FAM173B|family with sequence similarity 173, member B | -122 (-) | CCT5|chaperonin containing TCP1, subunit 5 (epsilon) | 198 (+) | -87:0.217 |
|
| chr19:1344799..1344971 | 18.359 | EFNA2|ephrin-A2 | -58920 (+) | MUM1|melanoma associated antigen (mutated) 1 | 10116 (+) | ||
| chr5:150508429..150508577 | 18.337 | TNIP1|TNFAIP3 interacting protein 1 | -41283 (-) | ANXA6|annexin A6 | 12704 (-) | 12:0.313, 13:0.313, 14:0.313 |
|
| chr4:174291838..174291976 | 18.297 | HMGB2|high mobility group box 2 | -36378 (-) | SAP30|Sin3A-associated protein, 30kDa | 156 (+) | ||
| chr6:16317039..16317161 | 18.224 | GMPR|guanosine monophosphate reductase | -78249 (+) | ATXN1|ataxin 1 | 444597 (-) | 2:0.846, 17:0.851, -99:0.955 |
-27:0.441 |
| chrX:115074642..115074746 | 18.187 | PLS3|plastin 3 | -246826 (+) | LOC642776|uncharacterized LOC642776 | 10745 (-) | ||
| chr7:129650819..129650998 | 18.185 | UBE2H|ubiquitin-conjugating enzyme E2H | -58099 (-) | ZC3HC1|zinc finger, C3HC-type containing 1 | 40313 (-) | 29:0.644, 30:0.289, 44:0.593, 45:0.272 |
|
| chr17:59940914..59941085 | 18.138 | BRIP1|BRCA1 interacting protein C-terminal helicase 1 | -205 (-) | INTS2|integrator complex subunit 2 | 64343 (-) | 25:0.320, 26:0.320, 27:0.320 |
|
| chr12:57151398..57151524 | 18.128 | PRIM1|primase, DNA, polypeptide 1 (49kDa) | -5335 (-) | HSD17B6|hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) | 5650 (+) | ||
| chr19:17685043..17685183 | 18.115 | GLT25D1|glycosyltransferase 25 domain containing 1 | -18602 (+) | UNC13A|unc-13 homolog A (C. elegans) | 113985 (-) | 36:0.841 |
-19:0.939 |
| chrX:130896839..130896960 | 18.049 | OR13H1|olfactory receptor, family 13, subfamily H, member 1 | -218901 (+) | LOC286467|family with sequence similarity 195, member A pseudogene | 67752 (-) | ||
| chr5:169233143..169233284 | 18.036 | DOCK2|dedicator of cytokinesis 2 | -168913 (+) | FAM196B|family with sequence similarity 196, member B | 174530 (-) | ||
| chr6:39948962..39949106 | 17.995 | MOCS1|molybdenum cofactor synthesis 1 | -46820 (-) | LRFN2|leucine rich repeat and fibronectin type III domain containing 2 | 606110 (-) | -35:0.882 |
|
| chr12:117295924..117296040 | 17.909 | RNFT2|ring finger protein, transmembrane 2 | -119863 (+) | HRK|harakiri, BCL2 interacting protein (contains only BH3 domain) | 23249 (-) | 21:0.747, -48:0.238, -49:0.238, -50:0.238, -51:0.243 |
|
| chr1:32986163..32986291 | 17.902 | ZBTB8B|zinc finger and BTB domain containing 8B | -55575 (+) | ZBTB8A|zinc finger and BTB domain containing 8A | 18544 (+) | -22:0.218 |
|
| chr4:75055126..75055262 | 17.841 | MTHFD2L|methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like | -31329 (+) | EPGN|epithelial mitogen homolog (mouse) | 119025 (+) | -25:0.796 |
|
| chr3:47018449..47018562 | 17.829 | CCDC12|coiled-coil domain containing 12 | -260 (-) | NBEAL2|neurobeachin-like 2 | 2742 (+) | -40:0.414, -48:0.311 |
|
| chr11:7716017..7716145 | 17.816 | CYB5R2|cytochrome b5 reductase 2 | -21329 (-) | OVCH2|ovochymase 2 (gene/pseudogene) | 11859 (-) | 0:0.996 |
|
| chr12:34175099..34175245 | 17.807 | SYT10|synaptotagmin X | -582506 (-) | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | 43 (+) | 20:0.238, 21:0.672, 36:0.723 |
-38:0.912 |
| chr4:99850348..99850528 | 17.787 | EIF4E|eukaryotic translation initiation factor 4E | -147 (-) | EIF4E|eukaryotic translation initiation factor 4E | 1348 (-) | ||
| chr12:923484..923594 | 17.763 | WNK1|WNK lysine deficient protein kinase 1 | -60783 (+) | WNK1|WNK lysine deficient protein kinase 1 | 93543 (+) | -43:0.817 |
6:0.398 |
| chr16:31884931..31885108 | 17.756 | ZNF720|zinc finger protein 720 | -160418 (+) | ZNF267|zinc finger protein 267 | 68 (+) | -32:0.238, -33:0.238, -34:0.238, -35:0.238 |
|
| chr9:139965039..139965190 | 17.749 | C9orf140|chromosome 9 open reading frame 140 | -81 (-) | UAP1L1|UDP-N-acteylglucosamine pyrophosphorylase 1-like 1 | 6839 (+) | 13:0.458, 14:0.458 |
|
| chr12:51318262..51318384 | 17.746 | TMPRSS12|transmembrane (C-terminal) protease, serine 12 | -81600 (+) | METTL7A|methyltransferase like 7A | 201 (+) | -16:0.778 |
|
| chr19:1194671..1194851 | 17.737 | SBNO2|strawberry notch homolog 2 (Drosophila) | -20482 (-) | STK11|serine/threonine kinase 11 | 11045 (+) | -83:0.955 |
|
| chrX:56829350..56829483 | 17.728 | LOC550643|uncharacterized LOC550643 | -73725 (+) | SPIN3|spindlin family, member 3 | 192555 (-) | -44:0.251 |
|
| chr8:72740225..72740387 | 17.703 | -280397 (-) | LOC100132891|uncharacterized LOC100132891 | 15968 (+) | 16:0.794 |
||
| chr2:102268294..102268474 | 17.664 | RFX8|regulatory factor X, 8 | -177191 (-) | MAP4K4|mitogen-activated protein kinase kinase kinase kinase 4 | 45968 (+) | -22:0.250, -23:0.674, -38:0.821 |
|
| chr6:87776274..87776415 | 17.608 | HTR1E|5-hydroxytryptamine (serotonin) receptor 1E | -129321 (+) | CGA|glycoprotein hormones, alpha polypeptide | 28474 (-) | -40:0.295, -41:0.295, -42:0.295 |
-6:0.404 |
| chr12:50419377..50419527 | 17.603 | RACGAP1|Rac GTPase activating protein 1 | -219 (-) | ACCN2|amiloride-sensitive cation channel 2, neuronal | 32109 (+) | 2:0.699 |
|
| chr17:5100414..5100537 | 17.579 | LOC100130950|uncharacterized LOC100130950 | -5095 (+) | SCIMP|SLP adaptor and CSK interacting membrane protein | 37626 (-) | -32:0.984 |
|
| chr19:12833368..12833526 | 17.577 | TNPO2|transportin 2 | -299 (-) | TNPO2|transportin 2 | 36 (-) | -28:0.399, -33:0.497, -49:0.846, -64:0.853 |
|
| chr20:3096315..3096431 | 17.567 | AVP|arginine vasopressin | -31021 (-) | FASTKD5|FAST kinase domains 5 UBOX5|U-box domain containing 5 | 44115 (-) | ||
| chr14:93700449..93700563 | 17.557 | UBR7|ubiquitin protein ligase E3 component n-recognin 7 (putative) | -26872 (+) | BTBD7|BTB (POZ) domain containing 7 | 98859 (-) | ||
| chr14:106144043..106144170 | 17.556 | IGHG2|immunoglobulin heavy constant gamma 2 (G2m marker) | -32990 (-) | IGHA1|immunoglobulin heavy constant alpha 1 | 30847 (-) | 44:0.494, 45:0.494 |
|
| chr6:20528673..20528819 | 17.535 | E2F3|E2F transcription factor 3 | -124713 (+) | CDKAL1|CDK5 regulatory subunit associated protein 1-like 1 | 5977 (+) | 7:0.665 |
|
| chr3:27722779..27722926 | 17.448 | SLC4A7|solute carrier family 4, sodium bicarbonate cotransporter, member 7 | -196922 (-) | EOMES|eomesodermin | 40941 (-) | 13:0.972 |
|
| chr6:38670998..38671156 | 17.445 | GLO1|glyoxalase I | -140 (-) | DNAH8|dynein, axonemal, heavy chain 8 | 12039 (+) | 24:0.330, 25:0.330, 26:0.330 |
|
| chr14:76127390..76127547 | 17.408 | C14orf1|chromosome 14 open reading frame 1 | -272 (-) | C14orf1|chromosome 14 open reading frame 1 | 87 (-) | 0:0.435, 1:0.435 |
|
| chr22:20772536..20772657 | 17.405 | ZNF74|zinc finger protein 74 | -24151 (+) | SCARF2|scavenger receptor class F, member 2 | 19466 (-) | 5:0.983 |
|
| chrX:134099531..134099658 | 17.390 | MOSPD1|motile sperm domain containing 1 | -50322 (-) | CXorf69|chromosome X open reading frame 69 | 25373 (+) | 31:0.527, 32:0.470 |
-16:0.776 |
| chr19:10369751..10369900 | 17.359 | MRPL4|mitochondrial ribosomal protein L4 | -6833 (+) | ICAM1|intercellular adhesion molecule 1 | 11691 (+) | -30:0.256, -45:0.246 |
28:0.961 |
| chr11:3585778..3585890 | 17.321 | ZNF195|zinc finger protein 195 | -185455 (-) | ART5|ADP-ribosyltransferase 5 | 77680 (-) | ||
| chr18:3593940..3594119 | 17.287 | TGIF1|TGFB-induced factor homeobox 1 | -140258 (+) | FLJ35776|uncharacterized LOC649446 | 113 (+) | -11:0.425, -12:0.425 |
|
| chr6:97731155..97731313 | 17.282 | MMS22L|MMS22-like, DNA repair protein | -144 (-) | POU3F2|POU class 3 homeobox 2 | 1551346 (+) | 39:0.694 |
|
| chr19:46898909..46899020 | 17.272 | PPP5C|protein phosphatase 5, catalytic subunit | -48605 (+) | CCDC8|coiled-coil domain containing 8 | 17907 (-) | -53:0.543, -68:0.533 |
50:0.996 |
| chr11:118955805..118955980 | 17.269 | HMBS|hydroxymethylbilane synthase | -237 (+) | HMBS|hydroxymethylbilane synthase | 2818 (+) | -2:0.713, -3:0.275, 12:0.849 |
|
| chr8:71520546..71520688 | 17.264 | TRAM1|translocation associated membrane protein 1 | -95 (-) | TRAM1|translocation associated membrane protein 1 | 39 (-) | 0:0.224, 0:0.361 |
|
| chr2:233470601..233470754 | 17.258 | EIF4E2|eukaryotic translation initiation factor 4E family member 2 | -55232 (+) | EFHD1|EF-hand domain family, member D1 | 89 (+) | 17:0.964 |
|
| chr12:1058844..1058998 | 17.238 | RAD52|RAD52 homolog (S. cerevisiae) | -95 (-) | ERC1|ELKS/RAB6-interacting/CAST family member 1 | 41531 (+) | 7:0.205, 8:0.207, 9:0.315 |
|
| chr12:49147365..49147480 | 17.233 | CCNT1|cyclin T1 | -36804 (-) | ADCY6|adenylate cyclase 6 | 30471 (-) | 50:0.818, 65:0.389, 66:0.422 |
12:0.982 |
| chrX:56790234..56790366 | 17.176 | LOC550643|uncharacterized LOC550643 | -34609 (+) | SPIN3|spindlin family, member 3 | 231671 (-) | -40:0.698 |
70:0.361 |
| chr8:7305809..7305940 | 17.139 | DEFB103A|defensin, beta 103A DEFB103B|defensin, beta 103B | -18005 (-) | SPAG11B|sperm associated antigen 11B | 4012 (-) | -26:0.955, -41:0.973 |
|
| chr3:127309593..127309754 | 17.138 | TPRA1|transmembrane protein, adipocyte asscociated 1 | -140 (-) | MCM2|minichromosome maintenance complex component 2 | 7567 (+) | -1:0.714, -2:0.277 |
|
| chr15:75746488..75746649 | 17.133 | SIN3A|SIN3 transcription regulator homolog A (yeast) | -2511 (-) | SIN3A|SIN3 transcription regulator homolog A (yeast) | 1555 (-) | ||
| chr11:124543697..124543832 | 17.119 | SIAE|sialic acid acetylesterase | -77 (-) | SPA17|sperm autoantigenic protein 17 | 11 (+) | 5:0.848 |
|
| chr18:12883796..12883937 | 17.116 | PSMG2|proteasome (prosome, macropain) assembly chaperone 2 | -180830 (+) | PTPN2|protein tyrosine phosphatase, non-receptor type 2 | 370 (-) | ||
| chr11:133280439..133280614 | 17.110 | OPCML|opioid binding protein/cell adhesion molecule-like | -466921 (-) | OPCML|opioid binding protein/cell adhesion molecule-like | 121788 (-) | 0:0.320, 2:0.411, 17:0.218, -32:0.777, 34:0.215, 35:0.282 |
|
| chr1:228594520..228594643 | 17.098 | TRIM11|tripartite motif containing 11 | -79 (-) | TRIM17|tripartite motif containing 17 | 9800 (-) | -3:0.221, 18:0.221 |
|
| chr17:7816401..7816525 | 17.097 | CHD3|chromodomain helicase DNA binding protein 3 | -3703 (+) | KCNAB3|potassium voltage-gated channel, shaker-related subfamily, beta member 3 | 16289 (-) | ||
| chr17:66031619..66031789 | 17.074 | C17orf58|chromosome 17 open reading frame 58 | -42031 (-) | KPNA2|karyopherin alpha 2 (RAG cohort 1, importin alpha 1) | 136 (+) | -14:0.499, -17:0.249 |
|
| chr15:44042151..44042312 | 17.072 | PDIA3|protein disulfide isomerase family A, member 3 | -3449 (+) | SERF2|small EDRK-rich factor 2 | 27074 (+) | 48:0.323, 49:0.340, 50:0.254, -60:0.421, -61:0.477 |
|
| chr8:42029311..42029437 | 17.070 | AP3M2|adaptor-related protein complex 3, mu 2 subunit | -18841 (+) | PLAT|plasminogen activator, tissue | 35714 (-) | ||
| chr3:101231971..101232143 | 17.048 | SENP7|SUMO1/sentrin specific peptidase 7 | -29 (-) | RG9MTD1|RNA (guanine-9-) methyltransferase domain containing 1 | 48660 (+) | 8:0.451, 9:0.309, 24:0.294, 25:0.435 |
|
| chr2:86595552..86595694 | 17.045 | REEP1|receptor accessory protein 1 | -30418 (-) | KDM3A|lysine (K)-specific demethylase 3A | 72647 (+) | 4:0.244, 6:0.493, 19:0.260, 21:0.525 |
|
| chr18:23747738..23747893 | 17.031 | PSMA8|proteasome (prosome, macropain) subunit, alpha type, 8 | -33967 (+) | TAF4B|TAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa | 58602 (+) | 1:0.238, 2:0.238, 3:0.238, 4:0.238 |
|
| chr16:68754104..68754242 | 16.995 | CDH3|cadherin 3, type 1, P-cadherin (placental) | -74919 (+) | CDH1|cadherin 1, type 1, E-cadherin (epithelial) | 17028 (+) | 2:0.906 |
|
| chr19:13305443..13305569 | 16.994 | IER2|immediate early response 2 | -41326 (+) | CACNA1A|calcium channel, voltage-dependent, P/Q type, alpha 1A subunit | 311670 (-) | -1:0.995 |
|
| chr12:97380862..97381002 | 16.948 | NEDD1|neural precursor cell expressed, developmentally down-regulated 1 | -79689 (+) | LOC643770|uncharacterized LOC643770 | 1516695 (-) | 24:0.412, 25:0.485, 39:0.227, 40:0.710 |
-15:0.919 |
| chr10:22629230..22629376 | 16.940 | BMI1|BMI1 polycomb ring finger oncogene | -19138 (+) | SPAG6|sperm associated antigen 6 | 5153 (+) | -21:0.760, 23:0.325 |
|
| chr1:51954061..51954228 | 16.923 | EPS15|epidermal growth factor receptor pathway substrate 15 | -66378 (-) | EPS15|epidermal growth factor receptor pathway substrate 15 | 30779 (-) | ||
| chr10:123494160..123494284 | 16.906 | FGFR2|fibroblast growth factor receptor 2 | -136464 (-) | ATE1|arginyltransferase 1 | 193303 (-) | -17:0.230, -18:0.230, -19:0.230, -20:0.230 |
12:0.992 |
| chr10:7914088..7914216 | 16.894 | TAF3|TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa | -53444 (+) | GATA3|GATA binding protein 3 | 182497 (+) | -47:0.326 |
-12:0.998 |
| chr1:200594608..200594777 | 16.887 | KIF14|kinesin family member 14 | -4813 (-) | DDX59|DEAD (Asp-Glu-Ala-Asp) box polypeptide 59 | 44337 (-) | -12:0.802, -27:0.791, 74:0.415, 75:0.415 |
|
| chr4:36843695..36843838 | 16.857 | DTHD1|death domain containing 1 | -558123 (+) | KIAA1239|KIAA1239 | 402923 (+) | ||
| chrX:48489909..48490044 | 16.857 | WDR13|WD repeat domain 13 | -33395 (+) | WAS|Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | 52198 (+) | ||
| chr18:39534909..39535061 | 16.854 | LOC647946|uncharacterized LOC647946 | -2154717 (-) | PIK3C3|phosphoinositide-3-kinase, class 3 | 214 (+) | -10:0.527, -11:0.470 |
|
| chr17:77720059..77720162 | 16.850 | ENPP7|ectonucleotide pyrophosphatase/phosphodiesterase 7 | -15221 (+) | CBX2|chromobox homolog 2 | 31871 (+) | -77:0.660, -78:0.251, -92:0.421, -93:0.447 |
14:0.264 |
| chr7:66949353..66949484 | 16.829 | STAG3L4|stromal antigen 3-like 4 | -181761 (+) | AUTS2|autism susceptibility candidate 2 | 2114418 (+) | -12:0.778 |
|
| chr7:76742172..76742309 | 16.807 | POMZP3|POM121 and ZP3 fusion | -485693 (-) | CCDC146|coiled-coil domain containing 146 | 9706 (+) | 24:0.996 |
|
| chr2:99453637..99453770 | 16.787 | MGAT4A|mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A | -106090 (-) | C2orf55|chromosome 2 open reading frame 55 | 98977 (-) | -23:0.234, -24:0.234, -25:0.234, -26:0.234 |
21:0.974 |
| chr7:22563688..22563807 | 16.781 | STEAP1B|STEAP family member 1B | -23900 (-) | IL6|interleukin 6 (interferon, beta 2) | 203076 (+) | 23:0.472, 24:0.472 |
-28:0.999 |
| chr11:67141092..67141234 | 16.780 | CLCF1|cardiotrophin-like cytokine factor 1 | -57 (-) | CLCF1|cardiotrophin-like cytokine factor 1 | 484 (-) | -6:0.765, -7:0.234, 29:0.783 |
|
| chr7:17022324..17022493 | 16.778 | AGR3|anterior gradient 3 homolog (Xenopus laevis) | -100804 (-) | AHR|aryl hydrocarbon receptor | 315861 (+) | 25:0.850 |
|
| chr12:112383174..112383281 | 16.775 | MAPKAPK5|mitogen-activated protein kinase-activated protein kinase 5 | -102907 (+) | LOC728543|uncharacterized LOC728543 TMEM116|transmembrane protein 116 | 67741 (-) | -55:0.854, -70:0.433, -71:0.518 |
-18:0.911 |
| chr2:74413126..74413306 | 16.739 | MOB1A|MOB kinase activator 1A | -7309 (-) | 12499 (-) | |||
| chr21:30848938..30849060 | 16.731 | BACH1|BTB and CNC homology 1, basic leucine zipper transcription factor 1 | -177263 (+) | GRIK1|glutamate receptor, ionotropic, kainate 1 | 463290 (-) | 29:0.330, 30:0.330, 31:0.330, -93:0.369 |
-50:0.951 |
| chr1:226548140..226548285 | 16.698 | LIN9|lin-9 homolog (C. elegans) | -50720 (-) | PARP1|poly (ADP-ribose) polymerase 1 | 47394 (-) | -21:0.940 |
|
| chr12:6667286..6667411 | 16.681 | IFFO1|intermediate filament family orphan 1 | -2090 (-) | NOP2|NOP2 nucleolar protein homolog (yeast) | 10118 (-) | 97:0.691 |
32:0.771 |
| chr11:63633791..63633930 | 16.670 | MARK2|MAP/microtubule affinity-regulating kinase 2 | -27256 (+) | MARK2|MAP/microtubule affinity-regulating kinase 2 | 22216 (+) | ||
| chr7:969736..969912 | 16.660 | ADAP1|ArfGAP with dual PH domains 1 | -9310 (-) | ADAP1|ArfGAP with dual PH domains 1 | 24291 (-) | ||
| chr3:50384725..50384872 | 16.650 | ZMYND10|zinc finger, MYND-type containing 10 | -1706 (-) | NPRL2|nitrogen permease regulator-like 2 (S. cerevisiae) | 3366 (-) | -15:0.596, -30:0.611 |
36:0.968 |
| chr5:64920474..64920635 | 16.618 | TRIM23|tripartite motif containing 23 | -387 (-) | C5orf44|chromosome 5 open reading frame 44 | 3 (+) | ||
| chr1:45860251..45860431 | 16.611 | TOE1|target of EGR1, member 1 (nuclear) | -54441 (+) | TESK2|testis-specific kinase 2 | 96492 (-) | 17:0.469, 18:0.359, 32:0.480, 33:0.438 |
|
| chr11:86002933..86003075 | 16.599 | EED|embryonic ectoderm development | -46718 (+) | C11orf73|chromosome 11 open reading frame 73 | 10365 (+) | -32:0.533, -47:0.380, -48:0.347 |
22:0.920 |
| chr20:25299252..25299411 | 16.597 | PYGB|phosphorylase, glycogen; brain | -70573 (+) | ABHD12|abhydrolase domain containing 12 | 72054 (-) | -22:0.419, -23:0.419 |
|
| chr6:161695077..161695224 | 16.591 | AGPAT4|1-acylglycerol-3-phosphate O-acyltransferase 4 (lysophosphatidic acid acyltransferase, delta) | -90 (-) | PACRG|PARK2 co-regulated | 1453019 (+) | -14:0.741 |
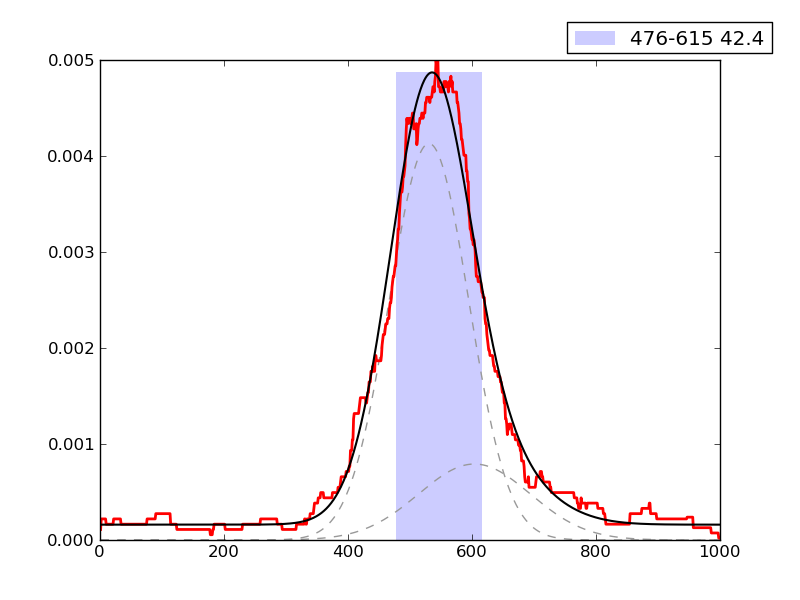
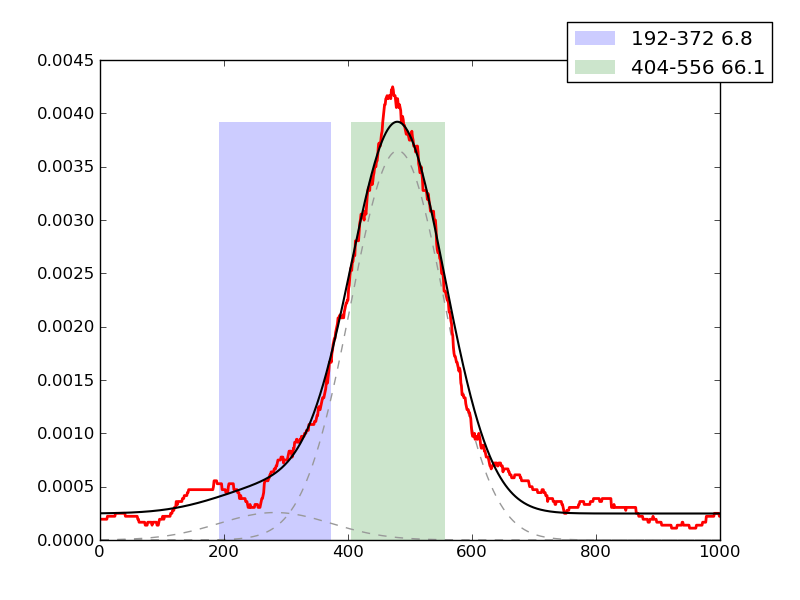
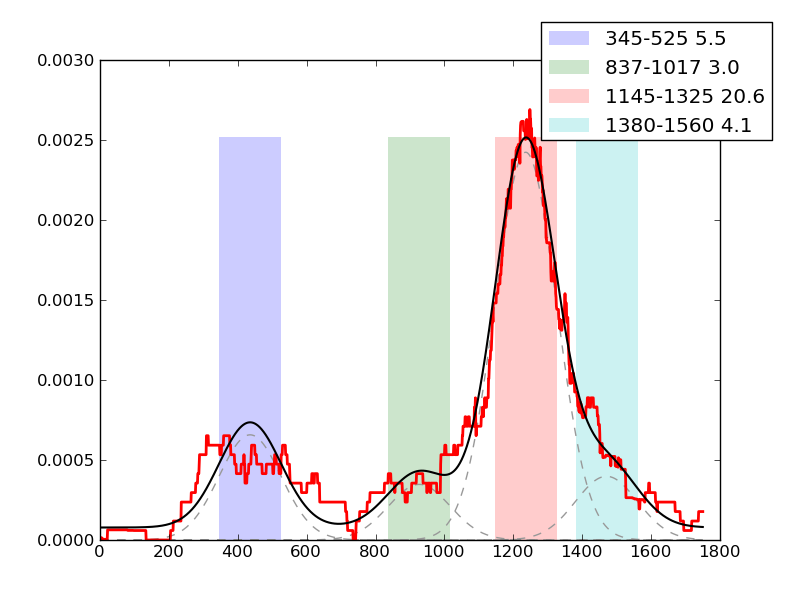
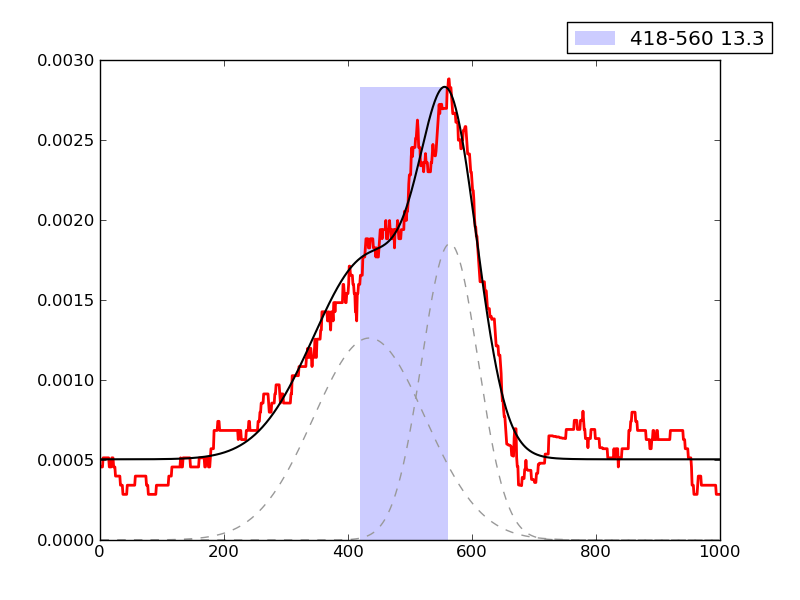
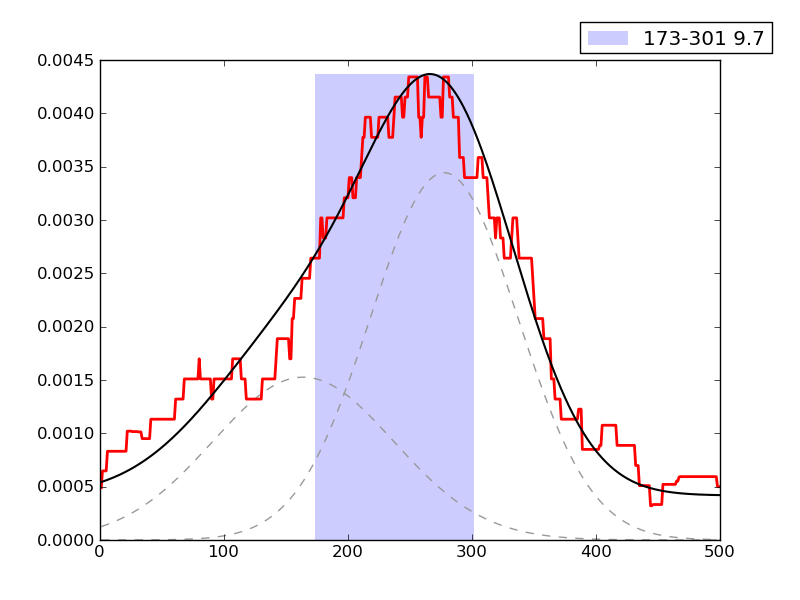
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
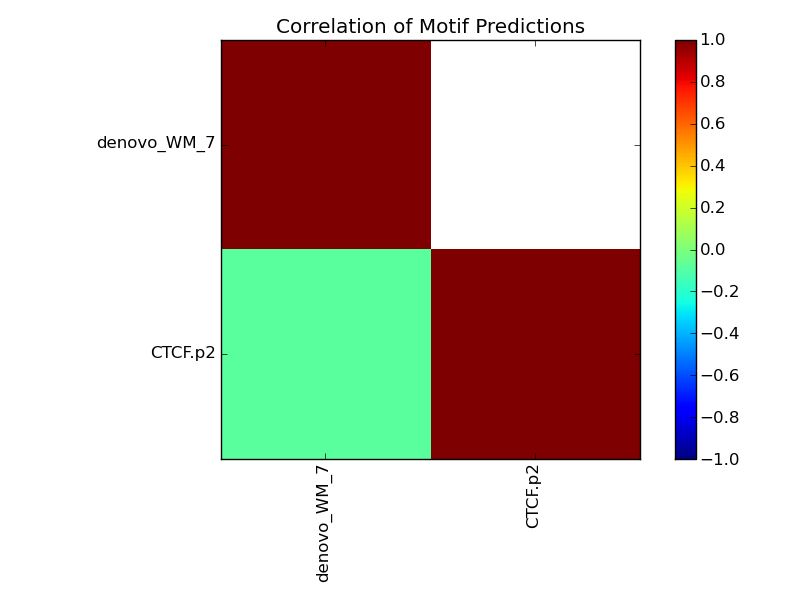
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| denovo_WM_7 | 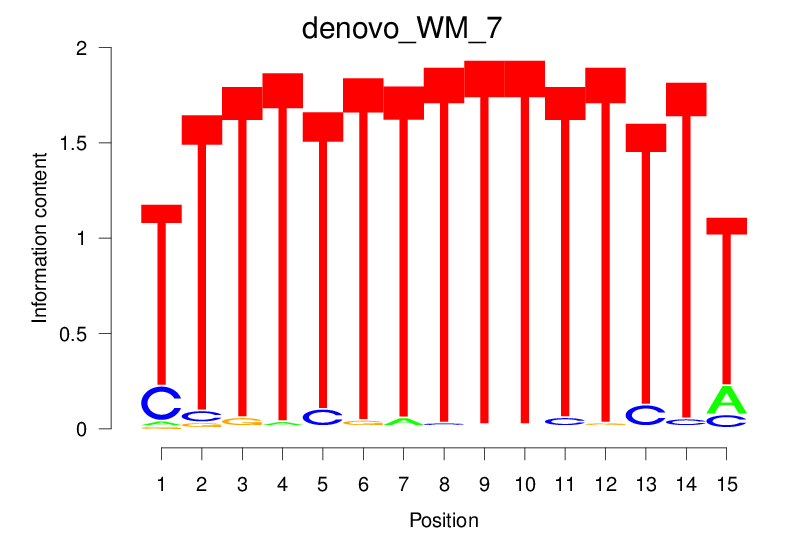 |
9.623 (1132.052) | 9.623 (1132.052) | 0.4637 | 0.1904 | 2.2838 | 12196/18267 |
| CTCF.p2 | 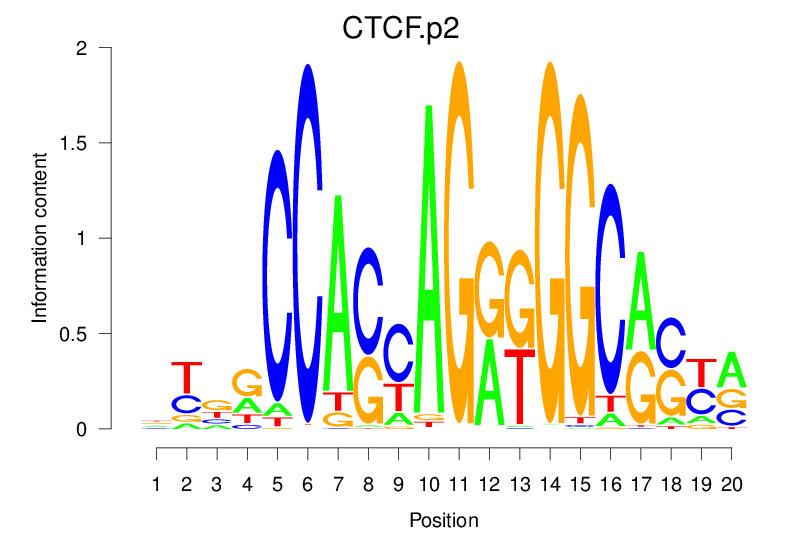 |
14.783 (1346.733) | 2.632 (483.81) | 0.3327 | 0.1255 | 4.3537 | 1517/18267 |
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| denovo_WM_7 |  |
9.623 (1132.052) | 0.4637 | 0.1904 | 2.2838 | 12196/18267 |
| denovo_WM_9 | 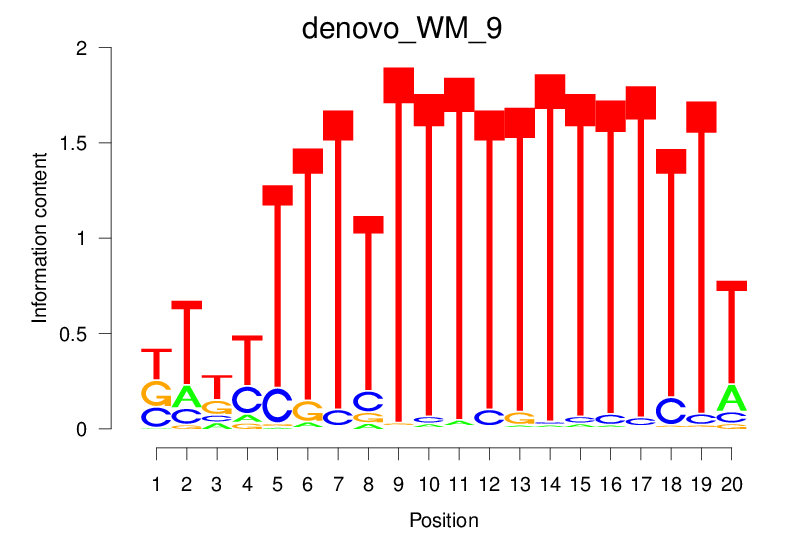 |
9.141 (1106.409) | 0.4619 | 0.1832 | 2.2131 | 12067/18267 |
| denovo_WM_17 | 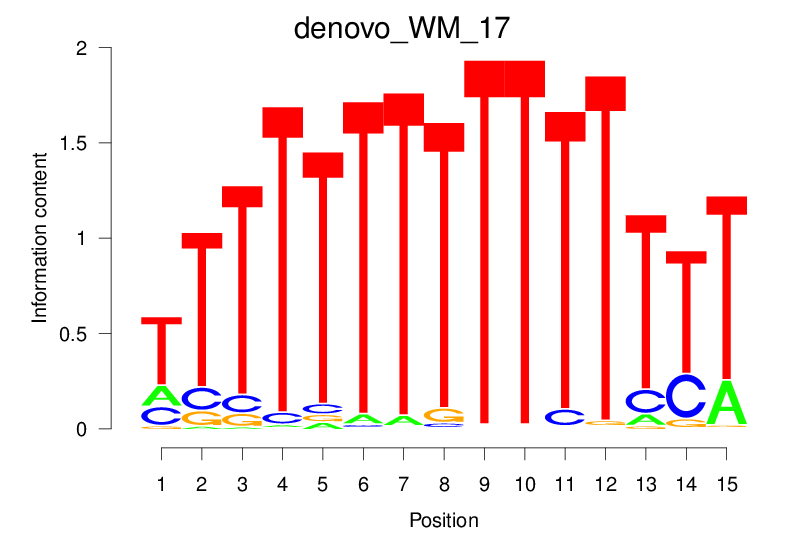 |
5.99 (895.016) | 0.5181 | 0.1817 | 2.0713 | 13539/18267 |
| denovo_WM_21 | 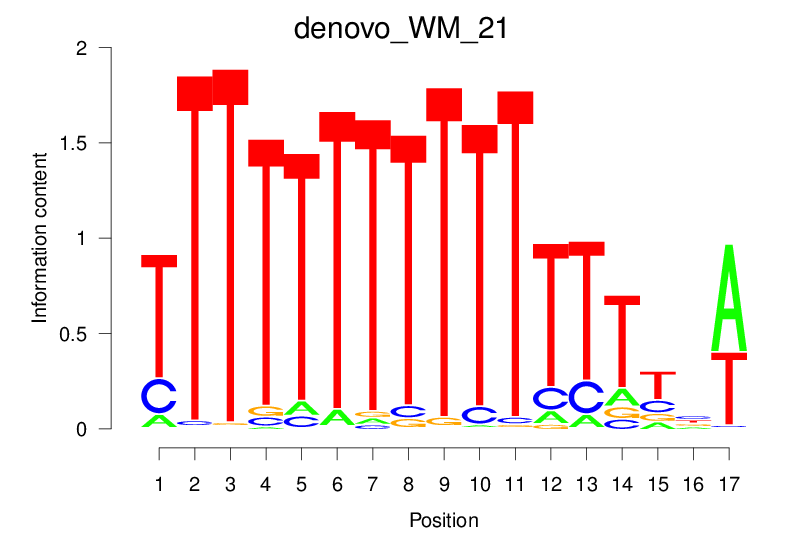 |
4.905 (795.173) | 0.5075 | 0.1782 | 2.0128 | 13673/18267 |
| denovo_WM_18 | 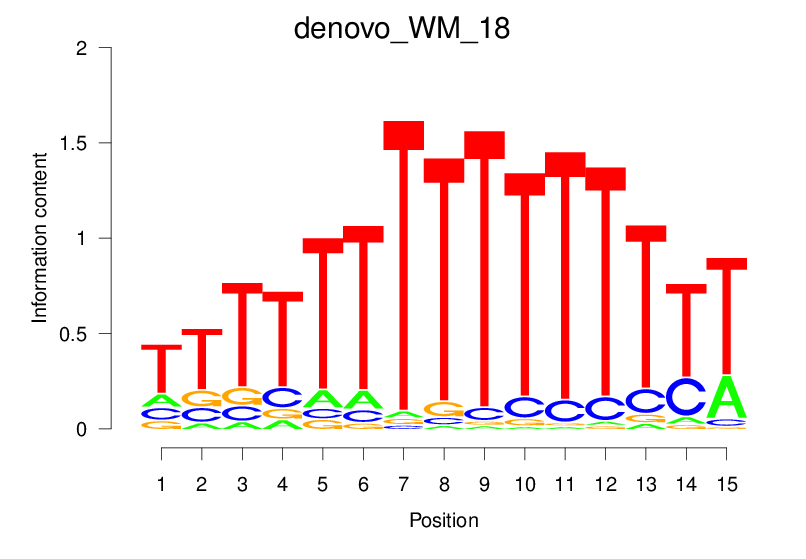 |
4.323 (731.962) | 0.5804 | 0.1639 | 1.846 | 15225/18267 |
| denovo_WM_8 | 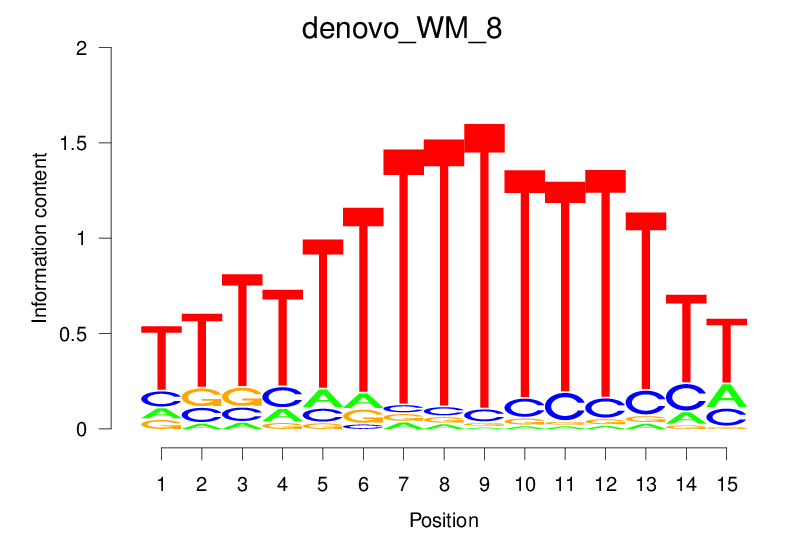 |
4.303 (729.628) | 0.5772 | 0.1642 | 1.8615 | 15167/18267 |
| denovo_WM_10 | 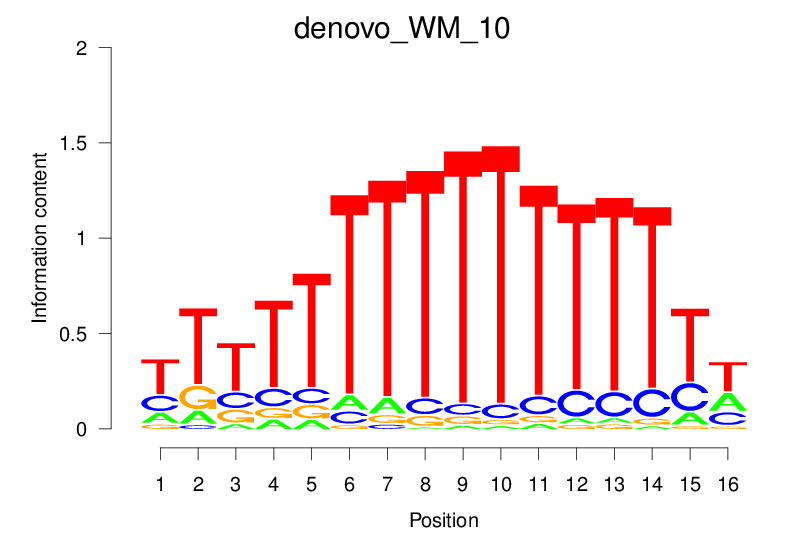 |
4.259 (724.572) | 0.5813 | 0.1628 | 1.813 | 15262/18267 |
| denovo_WM_22 | 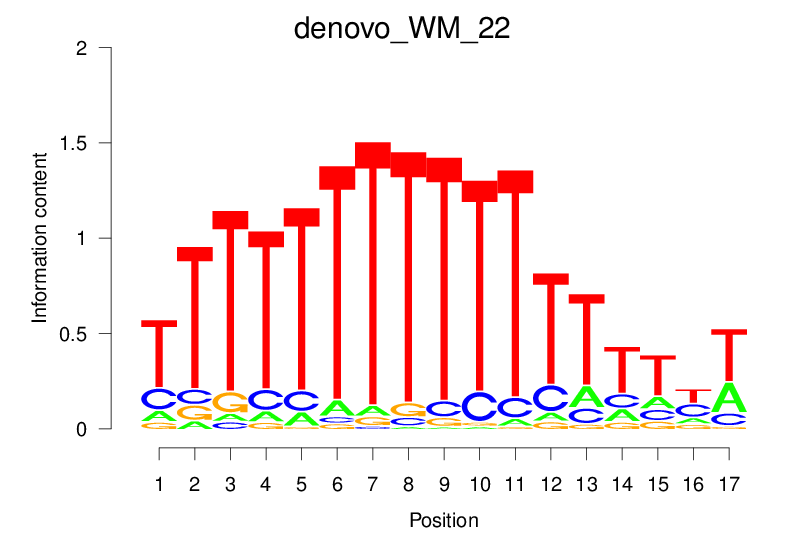 |
4.135 (709.713) | 0.5653 | 0.1589 | 1.8106 | 15123/18267 |
| HCMC.IRF4_si.wm | 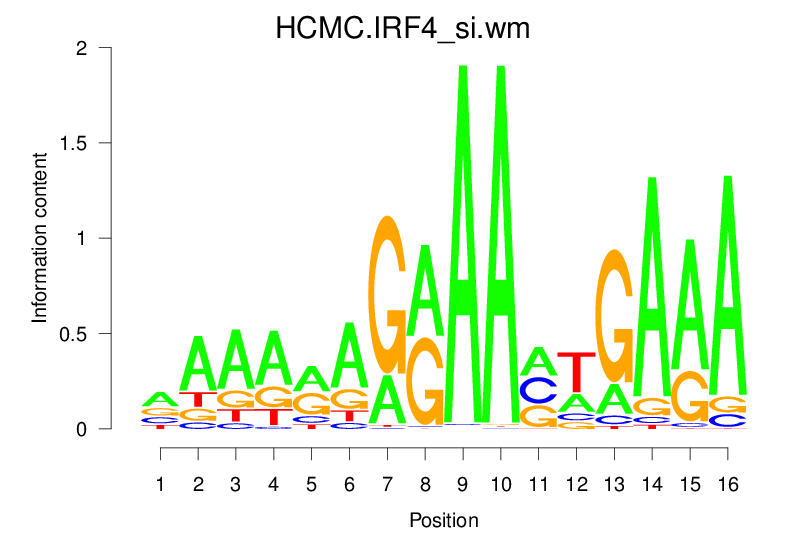 |
3.821 (670.294) | 0.5796 | 0.1697 | 1.9029 | 15127/18267 |
| denovo_WM_12 | 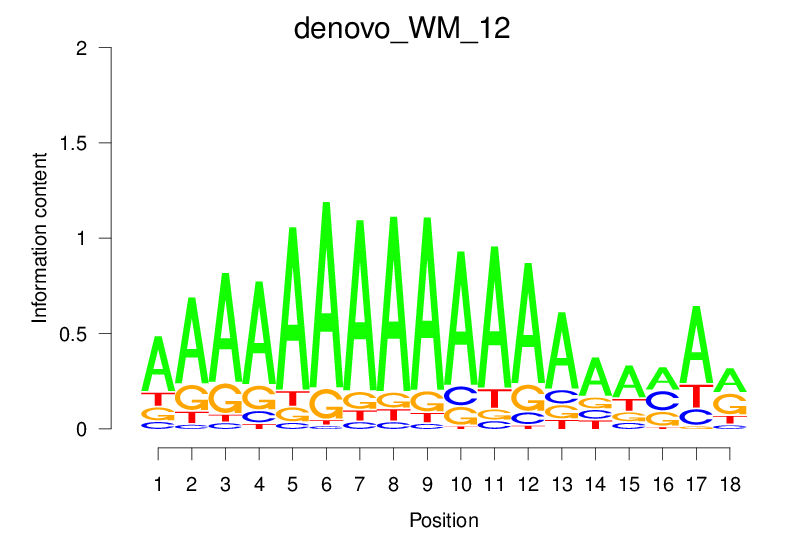 |
3.803 (667.839) | 0.5921 | 0.1466 | 1.6937 | 15841/18267 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis