CRUNCH Results
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.850 | 70th percentile | |
Fraction of mapped reads for the background samples: |
0.866 | 74th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.191 | 46th percentile | |
Error in fit of enrichment distribution: |
1.038 | 80th percentile | |
Peak statistics |
|||
Number of peaks: |
36955 | 81st percentile | |
Fraction of reads mapped to peaks: |
0.132 | 91st percentile | |
Motif enrichment statistics |
|||
Enrichment of top motif: |
40.87 | 87th percentile | |
Enrichment of complimentary motif set: |
61.79 | 89th percentile | |
Quality Control, Mapping, Fragment Size Estimation:
ZNF143 Replicate 1
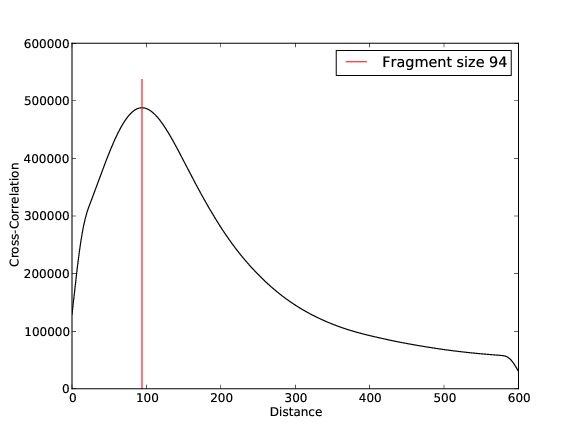
- 30,013,049 input reads (in FASTQ format).
- 27,745,544 reads after removal of low quality reads.
- 27,677,290 reads after removal of adapters and low complexity reads.
- 26,096,018 reads after mapping.
- Report PDF
ZNF143 Replicate 2
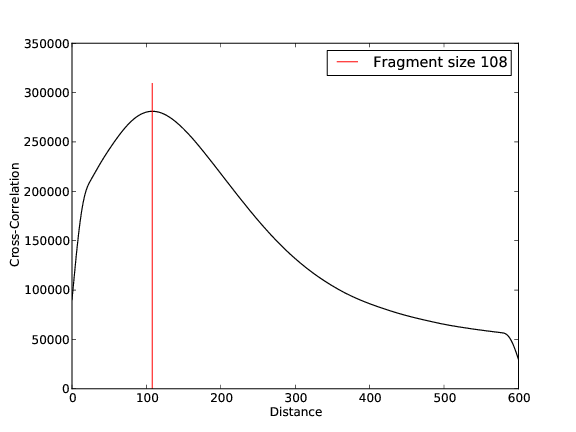
- 25,511,106 input reads (in FASTQ format).
- 23,462,173 reads after removal of low quality reads.
- 23,388,583 reads after removal of adapters and low complexity reads.
- 21,123,856 reads after mapping.
- Report PDF
BG Replicate 1
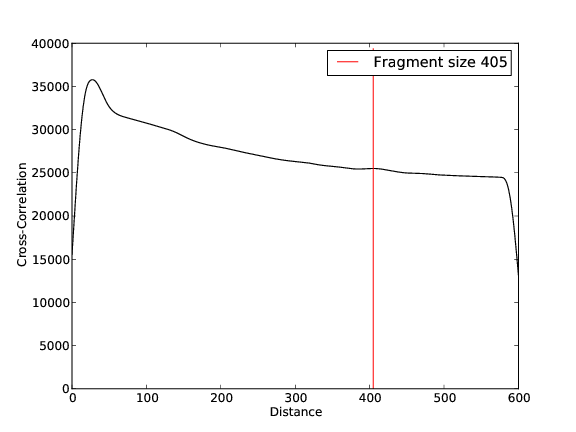
- 13,958,985 input reads (in FASTQ format).
- 13,816,004 reads after removal of low quality reads.
- 13,538,239 reads after removal of adapters and low complexity reads.
- 12,092,970 reads after mapping.
- Report PDF
Peak Calling:
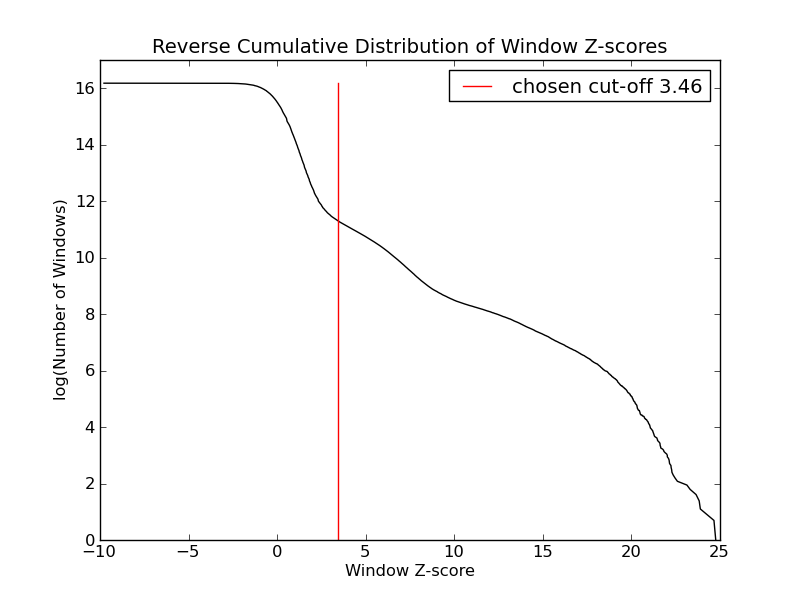
- 80008 windows are significantly enriched at FDR=10% (z-value cut-off: 3.46)
- 80008 windows merged to 40519 regions.
- 36955 significant peaks fitted within 40519 regions.
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) | denovo_WM_8 | ZNF143.p2 | denovo_WM_12 | STAT5{A,B}.p2 | denovo_WM_14 |
|---|---|---|---|---|---|---|---|---|---|---|
| chr19:54682363..54682447 | 24.337 | TMC4|transmembrane channel-like 4 | -5527 (-) | MBOAT7|membrane bound O-acyltransferase domain containing 7 | 9841 (-) | 30:1.000, -33:1.000 |
25:0.997, -29:0.978 |
34:0.963, -38:0.983 |
37:0.855, -40:0.864 |
|
| chr19:49310019..49310113 | 23.872 | FGF21|fibroblast growth factor 21 | -50648 (+) | BCAT2|branched chain amino-acid transaminase 2, mitochondrial | 4182 (-) | -28:1.000, 34:0.999 |
-23:0.997, 30:0.983 |
-32:0.980, 39:0.272 |
-35:0.864 |
|
| chr19:55815796..55815891 | 23.723 | BRSK1|BR serine/threonine kinase 1 | -2228 (+) | TMEM150B|transmembrane protein 150B | 20846 (-) | -31:0.727, 32:1.000 |
-27:0.489, 27:0.993 |
36:0.980 |
-38:0.917, 39:0.917 |
|
| chr19:49013917..49014022 | 23.464 | CYTH2|cytohesin 2 | -41300 (+) | 871 (-) | -25:1.000, 36:1.000 |
-20:0.999, 32:1.000 |
-29:0.980, 41:0.980 |
-32:0.917, 43:0.864 |
||
| chr7:150689938..150690041 | 22.892 | NOS3|nitric oxide synthase 3 (endothelial cell) | -1822 (+) | NOS3|nitric oxide synthase 3 (endothelial cell) | 875 (+) | -30:1.000, 31:1.000 |
27:0.808 |
-34:0.467, 36:0.908 |
35:0.893 |
-37:0.920, 38:0.511 |
| chr19:49148176..49148277 | 22.226 | DBP|D site of albumin promoter (albumin D-box) binding protein | -7622 (-) | CA11|carbonic anhydrase XI | 1125 (-) | 29:1.000, -33:1.000 |
25:0.989, -28:0.999 |
34:0.980, -37:0.980 |
36:0.920, -40:0.920 |
|
| chr17:47492318..47492458 | 22.223 | PHB|prohibitin | -140 (-) | NGFR|nerve growth factor receptor | 80300 (+) | -8:1.000, 79:0.998 |
-3:0.997, -70:0.874, 75:0.919 |
-12:0.980, 84:0.397 |
-15:0.920, 86:0.512 |
|
| chr19:47354267..47354386 | 22.210 | AP2S1|adaptor-related protein complex 2, sigma 1 subunit | -195 (-) | ARHGAP35|Rho GTPase activating protein 35 | 67606 (+) | 20:1.000, -22:1.000, -59:0.979, 75:1.000 |
15:0.999, -64:0.968, 70:0.998 |
24:0.968, -27:0.983, 79:0.270 |
24:0.933 |
27:0.855, -29:0.917 |
| chr3:69788436..69788556 | 22.173 | FRMD4B|FERM domain containing 4B | -196739 (-) | MITF|microphthalmia-associated transcription factor | 157 (+) | -24:1.000, 45:0.998 |
-28:0.996 |
-19:0.894, 41:0.662 |
-17:0.512, 38:0.657 |
|
| chr19:49250473..49250585 | 22.082 | IZUMO1|izumo sperm-egg fusion 1 | -434 (-) | FUT1|fucosyltransferase 1 (galactoside 2-alpha-L-fucosyltransferase, H blood group) | 8117 (-) | -29:1.000, 35:1.000 |
-25:1.000, 30:0.893 |
-34:0.980, 39:0.968 |
39:0.994 |
-36:0.857, 42:0.855 |
| chr13:41768687..41768802 | 22.014 | KBTBD7|kelch repeat and BTB (POZ) domain containing 7 | -119 (-) | MTRF1|mitochondrial translational release factor 1 | 68936 (-) | -34:0.960, 53:0.999 |
-5:0.458, -30:0.751, 48:0.921 |
57:0.241 |
||
| chr8:110346342..110346474 | 21.962 | NUDCD1|NudC domain containing 1 | -37 (-) | ENY2|enhancer of yellow 2 homolog (Drosophila) | 185 (+) | 7:0.999, -35:1.000, 65:0.449 |
-39:0.994, -64:0.885 |
-30:0.968 |
-31:0.663 |
14:0.855 |
| chr3:50329889..50330015 | 21.815 | IFRD2|interferon-related developmental regulator 2 | -89 (-) | HYAL3|hyaluronoglucosaminidase 3 NAT6|N-acetyltransferase 6 (GCN5-related) | 6339 (-) | -14:0.995, 40:0.999 |
35:0.971, 101:0.625 |
44:0.656 |
47:0.214 |
|
| chr20:62694359..62694479 | 21.720 | TCEA2|transcription elongation factor A (SII), 2 | -410 (+) | TCEA2|transcription elongation factor A (SII), 2 | 141 (+) | 15:0.998, -17:1.000, 75:0.963 |
-12:1.000, -61:0.365, 80:0.790 |
20:0.775, -21:0.931, 71:0.264 |
22:0.436, -24:0.702, 68:0.229 |
|
| chr12:110151305..110151415 | 21.627 | MVK|mevalonate kinase | -139729 (+) | C12orf34|chromosome 12 open reading frame 34 | 827 (+) | 24:1.000, -38:0.916 |
20:1.000 |
29:0.704 |
31:0.368, -45:0.917 |
|
| chr19:49362009..49362101 | 21.531 | HSD17B14|hydroxysteroid (17-beta) dehydrogenase 14 | -22132 (-) | PLEKHA4|pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 4 | 9741 (-) | 26:1.000, -35:0.966 |
22:0.883 |
31:0.803, -39:0.728 |
33:0.414, -42:0.917 |
|
| chr19:55816983..55817087 | 21.524 | BRSK1|BR serine/threonine kinase 1 | -3420 (+) | TMEM150B|transmembrane protein 150B | 19654 (-) | -30:1.000, 30:1.000 |
-25:0.998, 26:0.220 |
-34:0.936, 35:0.919 |
-37:0.920, 37:0.864 |
|
| chr19:49416518..49416625 | 21.502 | NUCB1|nucleobindin 1 | -12961 (+) | DHDH|dihydrodiol dehydrogenase (dimeric) | 20387 (+) | 29:1.000, -34:1.000 |
25:0.999, -29:0.974 |
34:0.980, -38:0.983 |
36:0.864, -41:0.920 |
|
| chr16:70473180..70473296 | 21.489 | ST3GAL2|ST3 beta-galactoside alpha-2,3-sialyltransferase 2 | -257 (-) | FUK|fucokinase | 15261 (+) | 9:1.000, 44:0.988, -49:1.000 |
14:0.999, -53:0.356 |
5:0.963, -44:0.306 |
5:0.878 |
2:0.849, -42:0.214, 51:0.864 |
| chr19:45988695..45988799 | 21.485 | FOSB|FBJ murine osteosarcoma viral oncogene homolog B | -17478 (+) | RTN2|reticulon 2 | 7746 (-) | -22:1.000, 40:1.000 |
-17:1.000, 36:0.998 |
-26:0.936, 45:0.968 |
-29:0.260, 47:0.849 |
|
| chr19:49248600..49248703 | 21.462 | RASIP1|Ras interacting protein 1 | -4745 (-) | IZUMO1|izumo sperm-egg fusion 1 | 1444 (-) | 30:1.000, -31:1.000 |
-26:0.393, 26:0.999 |
-35:0.728, 35:0.980 |
37:0.864, -38:0.864 |
|
| chr17:76210233..76210349 | 21.441 | AFMID|arylformamidase | -26868 (+) | BIRC5|baculoviral IAP repeat containing 5 | 8 (+) | -13:0.328, -45:0.999 |
-9:0.996, 34:0.961, -41:0.994 |
-50:0.980 |
-52:0.917 |
|
| chr12:54121309..54121460 | 21.422 | CALCOCO1|calcium binding and coiled-coil domain 1 | -130 (-) | HOXC13|homeobox C13 | 211248 (+) | 34:1.000, -36:0.998, -94:0.995, 95:0.888 |
29:1.000, -32:0.938, -99:0.886 |
38:0.609 |
41:0.238, -87:0.448 |
|
| chr17:7094941..7095032 | 21.387 | ASGR1|asialoglycoprotein receptor 1 | -12218 (-) | DLG4|discs, large homolog 4 (Drosophila) | 11608 (-) | 21:1.000, -35:0.999 |
17:0.999, -30:0.479 |
26:0.968, -39:0.808 |
25:0.344 |
28:0.849, -42:0.349 |
| chr19:49298763..49298876 | 21.349 | FGF21|fibroblast growth factor 21 | -39401 (+) | BCAT2|branched chain amino-acid transaminase 2, mitochondrial | 15429 (-) | 14:0.996, -48:1.000, 94:1.000 |
34:0.971, -44:0.973, 89:0.995 |
18:0.968, -53:0.980, 98:0.963 |
98:0.988 |
21:0.849, -55:0.365, 101:0.855 |
| chr19:48922662..48922771 | 21.336 | GRIN2D|glutamate receptor, ionotropic, N-methyl D-aspartate 2D | -24585 (+) | GRWD1|glutamate-rich WD repeat containing 1 | 26351 (+) | -32:1.000, 41:1.000 |
7:0.473, -28:0.999, -82:0.656 |
-37:0.968, 45:0.983 |
-36:0.682 |
-39:0.855, 48:0.920 |
| chr10:103113616..103113764 | 21.335 | LBX1|ladybird homeobox 1 | -124988 (-) | BTRC|beta-transducin repeat containing | 182 (+) | 0:0.926, 60:0.999, -85:1.000 |
-4:0.998 |
56:0.919, -89:0.983 |
53:0.920, -92:0.864 |
|
| chr20:62587826..62587942 | 21.156 | UCKL1|uridine-cytidine kinase 1-like 1 | -109 (-) | ZNF512B|zinc finger protein 512B | 13389 (-) | 0:0.999, 30:0.999, -31:0.999, 61:0.999, -62:0.999 |
3:0.974, -27:0.974, 34:0.974, -58:0.974, 65:0.974 |
-5:0.530, 25:0.530, -36:0.530, 56:0.530, -67:0.530, 93:0.254 |
-7:0.917, 23:0.917, -38:0.917, 54:0.917, -69:0.917 |
|
| chr19:49385135..49385230 | 21.132 | PPP1R15A|protein phosphatase 1, regulatory subunit 15A | -8693 (+) | TULP2|tubby like protein 2 | 16784 (-) | -31:1.000, 31:0.998 |
-27:0.697 |
35:0.963, -36:0.983 |
-38:0.864, 38:0.855 |
|
| chr1:2457649..2457765 | 20.996 | PLCH2|phospholipase C, eta 2 | -49950 (+) | PANK4|pantothenate kinase 4 | 357 (-) | -24:0.997, 34:0.992, -63:0.988, 70:0.992 |
-59:0.869 |
-17:0.645, 27:0.917, 63:0.917, -70:0.222 |
||
| chr18:71815636..71815776 | 20.989 | FBXO15|F-box protein 15 | -679 (-) | TIMM21|translocase of inner mitochondrial membrane 21 homolog (yeast) | 88 (+) | 30:0.982, -38:1.000 |
-43:0.950, 53:0.431 |
25:0.270, -34:0.958 |
-31:0.754 |
|
| chr12:49245955..49246078 | 20.941 | DDX23|DEAD (Asp-Glu-Ala-Asp) box polypeptide 23 | -84 (-) | RND1|Rho family GTPase 1 | 13584 (-) | 27:1.000, 76:0.957 |
32:0.653, -36:0.991, 72:0.843 |
23:0.274 |
||
| chr14:81408106..81408227 | 20.941 | CEP128|centrosomal protein 128kDa | -108 (-) | TSHR|thyroid stimulating hormone receptor | 13700 (+) | -26:1.000, 45:1.000 |
-31:0.990, 49:0.700 |
-22:0.980, 40:0.354 |
-19:0.917, 38:0.380 |
|
| chr19:47552051..47552158 | 20.814 | TMEM160|transmembrane protein 160 | -208 (-) | ZC3H4|zinc finger CCCH-type containing 4 | 63382 (-) | -16:0.995, 46:1.000 |
-11:0.736, 42:0.986 |
-20:0.604, 51:0.983 |
-23:0.581, 53:0.864 |
|
| chr20:32398923..32399061 | 20.805 | ZNF341|zinc finger protein 341 | -79196 (+) | CHMP4B|charged multivesicular body protein 4B | 94 (+) | 56:1.000, -86:1.000 |
0:0.477, 52:0.922, -81:1.000 |
61:0.980, -90:0.968 |
-90:0.907 |
63:0.302, -93:0.754 |
| chr16:30103244..30103360 | 20.768 | TBX6|T-box 6 | -55 (-) | YPEL3|yippee-like 3 (Drosophila) | 4243 (-) | 30:1.000, -32:1.000 |
26:1.000, -27:0.987 |
35:0.704, -36:0.931 |
-36:0.922 |
37:0.367, -39:0.713 |
| chr19:50651539..50651637 | 20.714 | ZNF473|zinc finger protein 473 | -122265 (+) | IZUMO2|IZUMO family member 2 | 14820 (-) | 28:1.000, -35:1.000 |
23:0.979, -31:0.968 |
32:0.691, -40:0.968 |
32:0.241, -39:0.871 |
-42:0.849 |
| chr1:247095485..247095601 | 20.679 | AHCTF1P1|AT hook containing transcription factor 1 pseudogene 1 AHCTF1|AT hook containing transcription factor 1 | -851 (-) | ZNF695|zinc finger protein 695 | 75769 (-) | -22:0.999, 41:0.541 |
-17:0.998 |
-26:0.662, 46:0.620 |
-29:0.432 |
|
| chr1:156737166..156737290 | 20.667 | HDGF|hepatoma-derived growth factor | -15097 (-) | PRCC|papillary renal cell carcinoma (translocation-associated) | 64 (+) | -16:0.997, 48:0.985 |
-12:0.998 |
-21:0.783, 52:0.301 |
-23:0.348, 55:0.920 |
|
| chr16:87984590..87984729 | 20.649 | CA5A|carbonic anhydrase VA, mitochondrial | -14512 (-) | BANP|BTG3 associated nuclear protein | 509 (+) | -15:1.000, 20:0.996, 60:1.000, -74:1.000 |
-10:0.956, 25:0.388, 56:1.000, -69:0.992 |
16:0.662, -19:0.936, 65:0.919, -78:0.936 |
13:0.581, -22:0.857, -81:0.313 |
|
| chr19:4791508..4791642 | 20.600 | MIR7-3HG|MIR7-3 host gene (non-protein coding) | -22414 (+) | FEM1A|fem-1 homolog a (C. elegans) | 161 (+) | 0:0.942, 51:0.993, -60:1.000 |
3:1.000, -65:0.909 |
55:0.931, -56:0.983 |
-53:0.917, 58:0.563 |
|
| chr10:38383147..38383271 | 20.591 | ZNF33A|zinc finger protein 33A | -83614 (+) | ZNF37A|zinc finger protein 37A | 74 (+) | 10:0.870, -35:1.000, 56:1.000 |
5:0.997, -40:0.997, 51:0.998 |
-31:0.968, 60:0.962 |
-28:0.754 |
|
| chr16:67840552..67840704 | 20.562 | RANBP10|RAN binding protein 10 | -119 (-) | TSNAXIP1|translin-associated factor X interacting protein 1 | 391 (+) | -20:0.968, 35:1.000 |
-15:0.310, 31:0.998 |
-36:0.680, 40:0.963 |
-27:0.223 |
|
| chr10:74870144..74870251 | 20.560 | P4HA1|prolyl 4-hydroxylase, alpha polypeptide I | -13427 (-) | NUDT13|nudix (nucleoside diphosphate linked moiety X)-type motif 13 | 54 (+) | 6:0.979, -22:1.000 |
1:0.957, -18:0.999 |
-27:0.983 |
-29:0.920, 70:0.236 |
|
| chr3:50397039..50397151 | 20.536 | TMEM115|transmembrane protein 115 | -157 (-) | CACNA2D2|calcium channel, voltage-dependent, alpha 2/delta subunit 2 | 143865 (-) | -5:1.000, 50:1.000, -68:0.999 |
-1:0.995, 45:1.000, -73:0.834 |
-10:0.936, 54:0.980 |
-12:0.920, 57:0.920 |
|
| chr1:54304239..54304366 | 20.533 | TMEM48|transmembrane protein 48 | -137 (-) | YIPF1|Yip1 domain family, member 1 | 51133 (-) | -1:0.999, 28:0.530, -56:1.000, 67:1.000 |
-5:0.859, 24:0.844, -60:1.000, 63:0.972 |
-51:0.908, 72:0.270 |
-49:0.499 |
|
| chr19:54024106..54024256 | 20.530 | ZNF813|zinc finger protein 813 | -53193 (+) | ZNF331|zinc finger protein 331 | 92 (+) | -29:1.000, 84:0.997 |
20:0.959, -24:0.979, -88:0.994, 89:0.999 |
-33:0.968 |
-33:0.321 |
-36:0.849 |
| chr1:45965776..45965921 | 20.525 | TESK2|testis-specific kinase 2 | -9015 (-) | MMACHC|methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria | 91 (+) | 9:0.996, -48:0.605, 68:0.818 |
14:0.958 |
5:0.264, 73:0.691, -100:0.503 |
2:0.238 |
|
| chr11:86013130..86013242 | 20.466 | EED|embryonic ectoderm development | -56900 (+) | C11orf73|chromosome 11 open reading frame 73 | 183 (+) | 18:0.976, -50:0.971 |
1:1.000 |
22:0.935 |
25:0.857 |
|
| chr11:65488277..65488371 | 20.458 | RNASEH2C|ribonuclease H2, subunit C | -49 (-) | DKFZp761E198|adaptor protein 5 | 59497 (-) | 26:0.999 |
0:0.883, 22:0.979 |
31:0.931 |
33:0.702 |
|
| chr11:16760039..16760170 | 20.437 | SOX6|SRY (sex determining region Y)-box 6 | -262179 (-) | C11orf58|chromosome 11 open reading frame 58 | 56 (+) | -31:1.000, 40:1.000 |
-35:0.999, 45:0.999 |
-26:0.983, 36:0.980 |
-24:0.864, 33:0.864 |
|
| chr6_cox_hap2:2387898..2388023 | 20.428 | None | None (None) | None | None (None) | -17:1.000, 41:1.000 |
45:1.000 |
-13:0.919, 36:0.809 |
-10:0.302, 34:0.227 |
|
| chr1:151138380..151138502 | 20.379 | LYSMD1|LysM, putative peptidoglycan-binding, domain containing 1 | -65 (-) | SCNM1|sodium channel modifier 1 | 121 (+) | -11:0.891, -52:0.999 |
-16:0.970, -48:0.999 |
-7:0.595, -57:0.450 |
-4:0.238 |
|
| chr10:103892641..103892765 | 20.364 | LDB1|LIM domain binding 1 | -12560 (-) | PPRC1|peroxisome proliferator-activated receptor gamma, coactivator-related 1 | 102 (+) | 16:0.999, 43:0.969, -54:0.997 |
12:0.964, 39:0.293, -49:0.981 |
21:0.596, 48:0.980, -58:0.241 |
23:0.427, 50:0.920 |
|
| chr21:15755496..15755623 | 20.348 | HSPA13|heat shock protein 70kDa family, member 13 | -86 (-) | SAMSN1|SAM domain, SH3 domain and nuclear localization signals 1 | 163090 (-) | -32:1.000, -56:0.239 |
-36:0.996 |
-27:0.968 |
-25:0.849 |
|
| chr9:111882268..111882395 | 20.340 | C9orf5|chromosome 9 open reading frame 5 | -111 (-) | C9orf4|chromosome 9 open reading frame 4 | 47198 (-) | 14:0.994, -43:0.998 |
-47:0.895 |
10:0.980, -38:0.980 |
7:0.864, -36:0.920 |
|
| chr6_mann_hap4:2223902..2224026 | 20.336 | None | None (None) | None | None (None) | -17:1.000, 41:1.000 |
45:1.000 |
-13:0.919, 36:0.809 |
-10:0.302, 34:0.227 |
|
| chr19:52097714..52097841 | 20.292 | ZNF175|zinc finger protein 175 | -23230 (+) | SIGLEC5|sialic acid binding Ig-like lectin 5 | 35880 (-) | 20:0.953, -46:0.982, 53:1.000 |
16:0.977, 49:0.978, -50:0.996, -93:0.321 |
-41:0.236, 58:0.868 |
60:0.581 |
|
| chr13:41706936..41707055 | 20.267 | KBTBD6|kelch repeat and BTB (POZ) domain containing 6 | -200 (-) | KBTBD7|kelch repeat and BTB (POZ) domain containing 7 | 61462 (-) | -33:0.986, 54:0.995 |
-3:0.726, -28:0.923, 50:0.478 |
-37:0.221, 59:0.241 |
||
| chr7:76751666..76751790 | 20.256 | POMZP3|POM121 and ZP3 fusion | -495181 (-) | CCDC146|coiled-coil domain containing 146 | 218 (+) | -29:0.984, 29:1.000 |
34:0.992 |
-24:0.446, 25:0.296 |
-22:0.427 |
|
| chr19:49947059..49947170 | 20.205 | SLC17A7|solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 | -2368 (-) | PIH1D1|PIH1 domain containing 1 | 7955 (-) | 20:1.000, -41:0.999 |
15:0.996, -37:0.957 |
24:0.963, -46:0.506 |
27:0.855 |
|
| chr1:6673616..6673738 | 20.201 | KLHL21|kelch-like 21 (Drosophila) | -10698 (-) | PHF13|PHD finger protein 13 | 109 (+) | 14:1.000, -45:0.996, 90:1.000 |
19:0.999, -49:0.991, 95:0.653 |
10:0.620, -40:0.219, 86:0.968 |
7:0.214, -38:0.349, 83:0.754 |
|
| chr14:73493922..73494063 | 20.191 | ZFYVE1|zinc finger, FYVE domain containing 1 | -161 (-) | RBM25|RNA binding motif protein 25 | 31217 (+) | 7:0.957, -61:0.999, 69:0.992 |
64:0.472, -66:1.000 |
2:0.980, 73:0.518 |
0:0.302, 76:0.236 |
|
| chr16:68482586..68482713 | 20.190 | SMPD3|sphingomyelin phosphodiesterase 3, neutral membrane (neutral sphingomyelinase II) | -168 (-) | ZFP90|zinc finger protein 90 homolog (mouse) | 81384 (+) | -31:1.000, 38:1.000, -66:0.979 |
-36:0.995, 42:0.999, 74:0.400 |
33:0.595 |
-27:0.374 |
31:0.229 |
| chr8:144897943..144898071 | 20.162 | SCRIB|scribbled homolog (Drosophila) | -463 (-) | PUF60|poly-U binding splicing factor 60KDa | 13200 (-) | -28:0.986 |
-23:0.997, 31:0.809, -78:0.220 |
|||
| chr19:54480853..54480960 | 20.158 | CACNG8|calcium channel, voltage-dependent, gamma subunit 8 | -14547 (+) | CACNG6|calcium channel, voltage-dependent, gamma subunit 6 | 14635 (+) | 29:1.000, -32:1.000 |
24:0.911, -28:0.999 |
33:0.968, -37:0.980 |
36:0.849, -39:0.857 |
|
| chr11:450193..450305 | 20.149 | ANO9|anoctamin 9 | -8246 (-) | PTDSS2|phosphatidylserine synthase 2 | 47 (+) | 0:0.998, 27:0.999 |
3:0.999, 31:0.997, -39:0.991 |
22:0.530 |
-7:0.436, 20:0.917 |
|
| chr19:48833418..48833530 | 20.142 | EMP3|epithelial membrane protein 3 | -4640 (+) | TMEM143|transmembrane protein 143 | 33795 (-) | -31:1.000, 31:0.999 |
-27:0.999 |
35:0.968, -36:0.775 |
-38:0.449, 38:0.754 |
|
| chr1:153931137..153931256 | 20.137 | CRTC2|CREB regulated transcription coactivator 2 | -79 (-) | SLC39A1|solute carrier family 39 (zinc transporter), member 1 | 4013 (-) | 7:1.000, -64:1.000, 84:0.983 |
-68:0.995, 80:0.872 |
3:0.499, -59:0.983 |
-57:0.864 |
|
| chr1:205744590..205744698 | 20.127 | RAB7L1|RAB7, member RAS oncogene family-like 1 | -66 (-) | SLC41A1|solute carrier family 41, member 1 | 37523 (-) | -26:0.999, 36:1.000 |
-22:0.998, 31:1.000 |
-31:0.968, 40:0.968 |
-33:0.226 |
|
| chr6_dbb_hap3:2169929..2170068 | 20.120 | None | None (None) | None | None (None) | -18:1.000, 40:1.000 |
44:1.000 |
-14:0.919, 35:0.809 |
-11:0.302, 33:0.227 |
|
| chr19:55690493..55690612 | 20.100 | C19orf51|chromosome 19 open reading frame 51 | -12569 (-) | SYT5|synaptotagmin V | 185 (-) | 26:1.000, -35:1.000 |
22:0.997, -30:0.933 |
31:0.980, -39:0.783 |
-42:0.360 |
|
| chr4:18023772..18023886 | 20.100 | LCORL|ligand dependent nuclear receptor corepressor-like | -363 (-) | SLIT2|slit homolog 2 (Drosophila) | 2231507 (+) | -27:0.983, 35:0.948 |
-23:0.901 |
-32:0.267 |
42:0.917 |
|
| chr19:44617384..44617534 | 20.097 | LOC100379224|uncharacterized LOC100379224 | -128 (-) | ZNF225|zinc finger protein 225 | 97 (+) | 17:1.000, -54:1.000, 73:0.999 |
12:1.000, -59:0.947, 68:0.810 |
21:0.931, -50:0.968, 77:0.499 |
21:0.975, -50:0.995, 77:0.983 |
24:0.702, -47:0.849 |
| chr19:49224336..49224452 | 20.090 | MAMSTR|MEF2 activating motif and SAP domain containing transcriptional regulator | -1428 (-) | RASIP1|Ras interacting protein 1 | 19512 (-) | -30:0.967, 31:0.999 |
26:0.964 |
-35:0.467, 35:0.301 |
-37:0.863, 38:0.920 |
|
| chr19:13885160..13885263 | 20.080 | MRI1|methylthioribose-1-phosphate isomerase homolog (S. cerevisiae) | -9820 (+) | C19orf53|chromosome 19 open reading frame 53 | 54 (+) | -69:0.386 |
-88:0.236 |
|||
| chr2:68694463..68694590 | 20.078 | FBXO48|F-box protein 48 | -116 (-) | APLF|aprataxin and PNKP like factor | 238 (+) | 28:0.994, -40:0.871 |
-35:0.274 |
21:0.917 |
||
| chr6:157801074..157801194 | 20.022 | TMEM242|transmembrane protein 242 | -55917 (-) | ZDHHC14|zinc finger, DHHC-type containing 14 | 1405 (+) | -25:0.999, 45:0.999 |
-30:0.999, -73:0.513 |
-21:0.936, 40:0.727 |
-18:0.864, 38:0.713 |
|
| chr1:26324602..26324739 | 20.015 | PAFAH2|platelet-activating factor acetylhydrolase 2, 40kDa | -80 (-) | EXTL1|exostoses (multiple)-like 1 | 23597 (+) | 1:0.974, 51:1.000, -69:0.584 |
56:0.999 |
-2:0.983, 47:0.920 |
-5:0.920, 44:0.576, -62:0.444 |
|
| chr6:30875829..30875942 | 20.011 | DDR1|discoidin domain receptor tyrosine kinase 1 | -19401 (+) | GTF2H4|general transcription factor IIH, polypeptide 4, 52kDa | 133 (+) | -17:1.000, 41:1.000 |
45:1.000 |
-13:0.919, 36:0.809 |
-10:0.302, 34:0.227 |
|
| chr19:48866418..48866527 | 19.996 | EMP3|epithelial membrane protein 3 | -37638 (+) | TMEM143|transmembrane protein 143 | 797 (-) | -28:1.000, 34:1.000 |
-24:0.937, 29:0.999 |
-33:0.980, 38:0.534 |
-35:0.920 |
|
| chr7:92861553..92861669 | 19.986 | HEPACAM2|HEPACAM family member 2 | -5828 (-) | CCDC132|coiled-coil domain containing 132 | 115 (+) | 20:1.000, -48:1.000 |
24:0.996, -53:0.689 |
15:0.908, -44:0.963 |
13:0.499, -41:0.849 |
|
| chr2:172543875..172543986 | 19.966 | CYBRD1|cytochrome b reductase 1 | -164975 (+) | DYNC1I2|dynein, cytoplasmic 1, intermediate chain 2 | 94 (+) | -6:1.000, 38:0.881, -41:0.999 |
-2:1.000, -37:0.996 |
-11:0.801, 42:0.467, -46:0.885 |
45:0.920 |
|
| chr16:11038278..11038398 | 19.965 | DEXI|Dexi homolog (mouse) | -2103 (-) | CLEC16A|C-type lectin domain family 16, member A | 105 (+) | 17:0.997, -40:0.999 |
21:0.993, -45:0.453 |
12:0.595 |
10:0.238, -33:0.917 |
|
| chr16:46918118..46918227 | 19.963 | C16orf87|chromosome 16 open reading frame 87 | -53084 (-) | GPT2|glutamic pyruvate transaminase (alanine aminotransferase) 2 | 109 (+) | -24:0.999, 34:0.998 |
-29:0.996, 38:0.921 |
-20:0.936 |
-17:0.920, 27:0.657 |
|
| chr17:7118403..7118504 | 19.947 | DLG4|discs, large homolog 4 (Drosophila) | -11859 (-) | DLG4|discs, large homolog 4 (Drosophila) | 2484 (-) | -31:0.985, 32:1.000 |
28:0.998 |
-35:0.775, 37:0.783 |
-38:0.449, 39:0.348 |
|
| chr19:49228236..49228332 | 19.928 | MAMSTR|MEF2 activating motif and SAP domain containing transcriptional regulator | -5318 (-) | RASIP1|Ras interacting protein 1 | 15622 (-) | -22:1.000, 39:1.000 |
-17:0.983, 35:0.996 |
-26:0.980, 44:0.980 |
43:0.203 |
-29:0.917, 46:0.917 |
| chr20:25677488..25677618 | 19.900 | ZNF337|zinc finger protein 337 | -194 (-) | LOC284801|uncharacterized LOC284801 | 512258 (-) | -10:0.997, -43:0.999, 59:0.999 |
-5:0.915, -38:0.982, 64:0.494 |
-47:0.387, 55:0.511 |
-47:0.958 |
-17:0.229, -50:0.702, 52:0.702 |
| chr1:156736965..156737129 | 19.889 | HDGF|hepatoma-derived growth factor | -14916 (-) | PRCC|papillary renal cell carcinoma (translocation-associated) | 245 (+) | -12:0.752, 36:0.981 |
41:0.530 |
43:0.302, -80:0.251 |
||
| chr16:12009917..12010059 | 19.887 | GSPT1|G1 to S phase transition 1 | -60 (-) | GSPT1|G1 to S phase transition 1 | 530 (-) | -12:0.927, 36:0.959, 81:0.850 |
31:0.588, 85:0.987 |
-19:0.920 |
||
| chr19:55709045..55709148 | 19.872 | SYT5|synaptotagmin V | -17503 (-) | PTPRH|protein tyrosine phosphatase, receptor type, H | 11768 (-) | 28:0.995, -36:1.000 |
24:0.223, -31:0.958 |
33:0.919, -40:0.908 |
35:0.917, -43:0.511 |
|
| chr14:97263510..97263625 | 19.865 | PAPOLA|poly(A) polymerase alpha | -294783 (+) | VRK1|vaccinia related kinase 1 | 148 (+) | 7:0.996, -25:1.000, -85:0.964 |
12:0.909, -20:1.000 |
3:0.374, -29:0.783, -89:0.680 |
-89:0.957 |
-32:0.227 |
| chr18:9475298..9475424 | 19.852 | TWSG1|twisted gastrulation homolog 1 (Drosophila) | -140504 (+) | RALBP1|ralA binding protein 1 | 236 (+) | 25:0.999, -45:1.000 |
29:0.999, -50:0.999 |
20:0.609, -41:0.696 |
18:0.238, -38:0.864 |
|
| chr17:7358804..7358920 | 19.844 | CHRNB1|cholinergic receptor, nicotinic, beta 1 (muscle) | -10012 (+) | ZBTB4|zinc finger and BTB domain containing 4 | 23991 (-) | 28:1.000, -35:0.997 |
23:0.978, -31:0.726 |
32:0.980, -40:0.908 |
-39:0.726 |
-42:0.657, -78:0.229 |
| chr1:156024459..156024589 | 19.841 | UBQLN4|ubiquilin 4 | -1009 (-) | LAMTOR2|late endosomal/lysosomal adaptor, MAPK and MTOR activator 2 | 73 (+) | -21:0.999, 46:0.998, -58:1.000 |
-17:0.994, 50:0.836, -54:1.000 |
-26:0.885, 41:0.980 |
-25:0.484 |
17:0.449, -28:0.593, 39:0.864 |
| chr10:124913601..124913714 | 19.820 | HMX2|H6 family homeobox 2 | -6020 (+) | BUB3|budding uninhibited by benzimidazoles 3 homolog (yeast) | 148 (+) | 23:0.998, -38:1.000 |
18:0.987, -34:1.000 |
27:0.920, -43:0.968 |
-42:0.994 |
30:0.576, -45:0.764 |
| chr19:49479584..49479692 | 19.819 | FTL|ferritin, light polypeptide | -10928 (+) | GYS1|glycogen synthase 1 (muscle) | 16804 (-) | 25:1.000, -34:0.997 |
21:1.000, -29:0.973 |
30:0.980 |
32:0.864, -82:0.296 |
|
| chr2:242448064..242448196 | 19.784 | STK25|serine/threonine kinase 25 | -99 (-) | BOK|BCL2-related ovarian killer | 50089 (+) | -14:1.000, 43:0.843, -63:0.226, -73:0.258, 82:0.999 |
-10:0.997, 38:0.971, -78:0.929, 86:0.998 |
-19:0.936 |
-21:0.917 |
|
| chr12:96429405..96429527 | 19.783 | LTA4H|leukotriene A4 hydrolase | -51 (-) | ELK3|ELK3, ETS-domain protein (SRF accessory protein 2) | 158660 (+) | 17:0.989, -40:0.987 |
21:0.984, -45:0.999 |
10:0.920 |
||
| chr17:73780702..73780824 | 19.776 | H3F3B|H3 histone, family 3B (H3.3B) | -4922 (-) | UNK|unkempt homolog (Drosophila) | 127 (+) | -6:0.945, -62:1.000, 66:1.000 |
-10:0.964, -57:0.992, 71:1.000 |
62:0.775, -66:0.735 |
-66:0.813 |
0:0.436, 59:0.449, -69:0.368 |
| chr12:32832065..32832182 | 19.769 | FGD4|FYVE, RhoGEF and PH domain containing 4 | -144860 (+) | DNM1L|dynamin 1-like | 34 (+) | -22:1.000, -64:0.817 |
-18:1.000, 23:0.994, -69:0.993 |
-27:0.980 |
-29:0.920 |
|
| chr19:44555991..44556129 | 19.734 | ZNF222|zinc finger protein 222 ZNF284|zinc finger protein 284 | -26538 (+) | ZNF223|zinc finger protein 223 | 122 (+) | 9:0.942, -59:0.848, -80:0.995, 80:0.987 |
5:0.833, 76:0.993, -84:0.990 |
-54:0.931 |
13:0.851, -76:0.395, 84:0.741 |
-52:0.702 |
| chr11:134123302..134123420 | 19.722 | THYN1|thymocyte nuclear protein 1 | -162 (-) | ACAD8|acyl-CoA dehydrogenase family, member 8 | 89 (+) | 25:1.000, -46:1.000 |
30:0.985, -50:0.998, -91:0.491 |
21:0.791, -41:0.983 |
-42:0.815 |
18:0.917, -39:0.917 |
| chr16:15149999..15150110 | 19.712 | NTAN1|N-terminal asparagine amidase | -177 (-) | RRN3|RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) | 38074 (-) | -14:1.000, 57:1.000, -90:0.979 |
-18:0.990, 53:0.993, -94:0.797 |
-9:0.968, 62:0.931, -85:0.499 |
-7:0.849, 64:0.702 |
|
| chr20:47805018..47805122 | 19.699 | STAU1|staufen, RNA binding protein, homolog 1 (Drosophila) | -157 (-) | DDX27|DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | 30822 (+) | 42:1.000, -73:0.705 |
-33:0.710, 37:0.999, -69:0.556 |
46:0.980 |
49:0.920, -80:0.449 |
|
| chr19:55629086..55629205 | 19.696 | PPP1R12C|protein phosphatase 1, regulatory subunit 12C | -213 (-) | TNNT1|troponin T type 1 (skeletal, slow) | 29272 (-) | -34:0.993, 101:1.000 |
30:0.576, -39:0.998, 96:1.000 |
-30:0.691 |
-27:0.214 |
|
| chr7:128864577..128864692 | 19.664 | SMO|smoothened, frizzled family receptor | -35922 (+) | AHCYL2|adenosylhomocysteinase-like 2 | 195 (+) | -31:1.000, 75:0.441 |
-26:0.978 |
-35:0.980 |
-38:0.917 |
|
| chr7:100888034..100888145 | 19.647 | CLDN15|claudin 15 | -5989 (-) | FIS1|fission 1 (mitochondrial outer membrane) homolog (S. cerevisiae) | 236 (-) | 31:0.999, -32:1.000 |
26:0.669, -28:1.000 |
35:0.534, -37:0.980 |
-39:0.239 |
|
| chr19:50145255..50145380 | 19.645 | RRAS|related RAS viral (r-ras) oncogene homolog | -1940 (-) | SCAF1|SR-related CTD-associated factor 1 | 113 (+) | 10:0.935, -39:0.999, 70:0.798 |
6:0.759, -34:0.998, 75:0.210 |
-43:0.609 |
-46:0.229 |
|
| chr4:2420543..2420652 | 19.642 | ZFYVE28|zinc finger, FYVE domain containing 28 | -193 (-) | LOC402160|uncharacterized LOC402160 | 100 (+) | 29:1.000, -44:1.000 |
25:0.993, -39:0.540 |
34:0.980, -48:0.936 |
36:0.917, -51:0.864 |
|
| chr2:217277055..217277184 | 19.621 | MARCH4|membrane-associated ring finger (C3HC4) 4 | -40412 (-) | SMARCAL1|SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a-like 1 | 79 (+) | -20:1.000, 64:1.000, -99:0.999 |
-15:0.990, 60:1.000, -94:0.992 |
-24:0.919, 69:0.968 |
-24:0.248 |
-27:0.920, 71:0.855 |
| chr17:17495034..17495168 | 19.613 | PEMT|phosphatidylethanolamine N-methyltransferase | -126 (-) | RAI1|retinoic acid induced 1 | 89684 (+) | -24:1.000, 57:0.995 |
-28:0.641, 62:0.256 |
-19:0.968, 53:0.936 |
-20:0.997 |
-17:0.855, 50:0.920 |
| chr12:133532873..133532977 | 19.604 | ZNF605|zinc finger protein 605 | -52 (-) | ZNF26|zinc finger protein 26 | 30073 (+) | 4:1.000, -27:1.000 |
8:0.913, -23:0.993 |
0:0.931, -32:0.968 |
-2:0.713, -34:0.855 |
|
| chr5:178286868..178286986 | 19.586 | ZNF354A|zinc finger protein 354A | -129259 (-) | ZNF354B|zinc finger protein 354B | 72 (+) | 15:0.974, -19:1.000 |
-15:0.395 |
10:0.691, -24:0.931 |
11:0.909, -23:0.994 |
8:0.214, -26:0.713 |
| chr19:16607030..16607163 | 19.571 | CALR3|calreticulin 3 | -126 (-) | C19orf44|chromosome 19 open reading frame 44 | 119 (+) | 18:0.844, -66:0.905, 81:1.000 |
-7:0.306, 22:0.941, 76:0.999 |
13:0.221, -71:0.980, 85:0.963 |
-73:0.864, 88:0.754 |
|
| chr11:72145626..72145736 | 19.536 | CLPB|ClpB caseinolytic peptidase B homolog (E. coli) | -31 (-) | PDE2A|phosphodiesterase 2A, cGMP-stimulated | 207798 (-) | 15:1.000, -42:0.938, 78:0.852 |
20:0.995, 74:0.412 |
11:0.803, -37:0.464 |
11:0.242 |
8:0.290 |
| chr7:150755072..150755214 | 19.532 | CDK5|cyclin-dependent kinase 5 | -128 (-) | SLC4A2|solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) | 155 (+) | -19:0.995, 64:0.999, -87:1.000 |
-14:0.828, 69:0.999, -91:0.999 |
-23:0.791, 60:0.282, -82:0.530 |
-26:0.857, -80:0.917 |
|
| chr19:48907000..48907108 | 19.531 | GRIN2D|glutamate receptor, ionotropic, N-methyl D-aspartate 2D | -8923 (+) | GRWD1|glutamate-rich WD repeat containing 1 | 42013 (+) | -20:1.000, 34:1.000 |
-24:0.999, 30:0.755 |
-15:0.963, 39:0.397 |
-13:0.754, 41:0.657 |
|
| chr2:39005271..39005379 | 19.523 | SRSF7|serine/arginine-rich splicing factor 7 | -26809 (-) | GEMIN6|gem (nuclear organelle) associated protein 6 | 30 (+) | -32:0.346 |
-35:0.917 |
|||
| chr5:173043722..173043845 | 19.517 | BOD1|biorientation of chromosomes in cell division 1 | -110 (-) | CPEB4|cytoplasmic polyadenylation element binding protein 4 | 271595 (+) | 0:1.000, 47:0.656 |
4:0.273, 43:0.980 |
-4:0.980, 52:0.236 |
-7:0.864 |
|
| chr11:43665688..43665800 | 19.510 | TTC17|tetratricopeptide repeat domain 17 | -285252 (+) | 40 (+) | 0:0.998, 24:0.993 |
-5:0.976, 19:0.859 |
3:0.604 |
|||
| chr18:77439373..77439541 | 19.508 | -144 (-) | FLJ25715|uncharacterized protein FLJ25715 | 284 (-) | -17:0.992, 55:0.999, -92:0.999, 96:0.999 |
-21:0.988, 51:0.995, -54:0.584, 92:0.975, -96:0.998 |
60:0.662, -87:0.254, 101:0.446 |
-10:0.432, 62:0.581 |
||
| chr19:56652287..56652426 | 19.489 | ZNF787|zinc finger protein 787 | -19710 (-) | ZNF444|zinc finger protein 444 | 249 (+) | -25:1.000, 69:0.999 |
-29:0.975 |
-20:0.968, 74:0.968 |
-21:0.979 |
-18:0.849, 76:0.855 |
| chr2:2617267..2617393 | 19.485 | MYT1L|myelin transcription factor 1-like | -282246 (-) | TSSC1|tumor suppressing subtransferable candidate 1 | 764226 (-) | 0:1.000, 59:0.992 |
5:1.000, -42:0.992, 64:0.999 |
-3:0.816 |
-6:0.864 |
|
| chr14:77923978..77924125 | 19.468 | C14orf133|chromosome 14 open reading frame 133 | -116 (-) | AHSA1|AHA1, activator of heat shock 90kDa protein ATPase homolog 1 (yeast) | 339 (+) | 15:0.998, -55:0.999 |
-59:1.000 |
11:0.397 |
-7:0.449, -48:0.444 |
|
| chr17:76250639..76250745 | 19.464 | TMEM235|transmembrane protein 235 | -23229 (+) | SOCS3|suppressor of cytokine signaling 3 | 105465 (-) | -29:1.000, 40:1.000 |
-34:1.000, 35:1.000 |
-25:0.980, 44:0.980 |
-22:0.920 |
|
| chr19:42748962..42749077 | 19.463 | GSK3A|glycogen synthase kinase 3 alpha | -2261 (-) | ERF|Ets2 repressor factor | 10195 (-) | 28:0.991, -56:1.000 |
24:0.981, -51:0.408 |
33:0.704, -60:0.586 |
-60:0.886 |
35:0.251, -63:0.754 |
| chr1:149871040..149871150 | 19.457 | HIST2H2AB|histone cluster 2, H2ab HIST2H2BE|histone cluster 2, H2be | -11582 (-) | BOLA1|bolA homolog 1 (E. coli) | 82 (+) | 30:1.000, -36:1.000 |
-31:0.524, 35:0.998 |
26:0.691, -40:0.908 |
23:0.214, -43:0.499, 60:0.229 |
|
| chr3:4344924..4345038 | 19.440 | LRRN1|leucine rich repeat neuronal 1 | -503882 (+) | SETMAR|SET domain and mariner transposase fusion gene | 27 (+) | 13:1.000, -19:1.000 |
-14:0.842, 18:1.000 |
9:0.506, -23:0.983 |
6:0.436, -26:0.920 |
|
| chr2:202316275..202316389 | 19.421 | TRAK2|trafficking protein, kinesin binding 2 | -44 (-) | STRADB|STE20-related kinase adaptor beta | 140 (+) | 20:1.000 |
-13:0.317, 15:0.994 |
24:0.983 |
27:0.864 |
|
| chr16:19535065..19535171 | 19.409 | GDE1|glycerophosphodiester phosphodiesterase 1 | -1742 (-) | CCP110|centriolar coiled coil protein 110kDa | 113 (+) | 27:0.913, -37:1.000 |
22:0.375, -33:0.867 |
-42:0.803 |
-44:0.279 |
|
| chr1:10532589..10532709 | 19.401 | DFFA|DNA fragmentation factor, 45kDa, alpha polypeptide | -27 (-) | PEX14|peroxisomal biogenesis factor 14 | 2362 (+) | 9:1.000, -41:1.000, 60:0.998 |
4:0.999, -46:0.991, 55:0.877 |
13:0.968, -37:0.963 |
16:0.855, -34:0.855, 67:0.499 |
|
| chrUn_gl000223:149688..149792 | 19.393 | None | None (None) | None | None (None) | -4:1.000, 27:1.000 |
-8:0.913, 23:0.993 |
0:0.931, 32:0.968 |
2:0.713, 34:0.855 |
|
| chr2:88991028..88991148 | 19.376 | EIF2AK3|eukaryotic translation initiation factor 2-alpha kinase 3 | -64110 (-) | RPIA|ribose 5-phosphate isomerase A | 127 (+) | 22:1.000, -36:0.999 |
26:0.974, -41:0.721 |
17:0.968, -32:0.274 |
18:0.592 |
15:0.855, -29:0.214 |
| chr8:124054093..124054203 | 19.361 | ZHX2|zinc fingers and homeoboxes 2 | -90449 (+) | DERL1|Der1-like domain family, member 1 | 359 (-) | 24:0.999, -38:1.000 |
20:0.947, -33:0.999 |
-42:0.919 |
31:0.499 |
|
| chr2:63815944..63816078 | 19.344 | WDPCP|WD repeat containing planar cell polarity effector | -164 (-) | MDH1|malate dehydrogenase 1, NAD (soluble) | 102 (+) | 27:1.000, -44:0.362 |
31:0.991 |
22:0.908, -40:0.931 |
23:0.392 |
20:0.511, -37:0.713 |
| chr19:1241637..1241756 | 19.342 | C19orf26|chromosome 19 open reading frame 26 | -3782 (-) | ATP5D|ATP synthase, H+ transporting, mitochondrial F1 complex, delta subunit | 56 (+) | 10:0.999, -45:0.994, 51:0.521 |
5:0.241, -41:0.983, 55:0.951 |
14:0.868 |
17:0.593, -52:0.251 |
|
| chr22:41682269..41682400 | 19.329 | RANGAP1|Ran GTPase activating protein 1 | -127 (-) | ZC3H7B|zinc finger CCCH-type containing 7B | 15180 (+) | 28:0.904, -31:0.675 |
32:0.918 |
|||
| chr12:88535944..88536074 | 19.326 | CEP290|centrosomal protein 290kDa | -140 (-) | TMTC3|transmembrane and tetratricopeptide repeat containing 3 | 139 (+) | -14:0.999, 23:1.000 |
-9:0.946, 19:0.910, 63:0.980 |
-18:0.234, 28:0.211 |
||
| chr5:145562231..145562378 | 19.323 | LARS|leucyl-tRNA synthetase | -40 (-) | RBM27|RNA binding motif protein 27 | 20813 (+) | 1:0.994, -57:1.000, 92:0.845 |
-62:0.899 |
-3:0.983, -53:0.983 |
-5:0.302, -50:0.920, 85:0.920 |
|
| chr19:51142549..51142651 | 19.320 | SYT3|synaptotagmin III | -1299 (-) | SYT3|synaptotagmin III | 525 (-) | -31:1.000, 31:0.989 |
26:0.981, -27:0.951 |
35:0.569, -36:0.983 |
-38:0.857 |
|
| chr6:5003887..5004003 | 19.317 | CDYL|chromodomain protein, Y-like | -113720 (+) | LOC285778|uncharacterized LOC285778 | 81 (+) | -27:1.000, 31:1.000 |
-31:0.999, 36:0.998 |
-22:0.980, 27:0.980 |
-20:0.916, 24:0.857 |
|
| chr5:40756036..40756144 | 19.309 | TTC33|tetratricopeptide repeat domain 33 | -50 (-) | PRKAA1|protein kinase, AMP-activated, alpha 1 catalytic subunit | 42189 (-) | 15:1.000, -26:0.999 |
11:0.781, -21:0.993 |
20:0.919, -30:0.443 |
-30:0.430 |
-33:0.436 |
| chr12:107167936..107168061 | 19.287 | RFX4|regulatory factor X, 4 (influences HLA class II expression) | -89465 (+) | RIC8B|resistance to inhibitors of cholinesterase 8 homolog B (C. elegans) | 464 (+) | -13:1.000, 52:0.998, -61:1.000 |
-9:0.964, 47:0.994, -57:0.998 |
-18:0.274, 56:0.735, -66:0.411 |
-65:0.270 |
59:0.368 |
| chr5:85913628..85913746 | 19.261 | EDIL3|EGF-like repeats and discoidin I-like domains 3 | -2233023 (-) | COX7C|cytochrome c oxidase subunit VIIc | 71 (+) | 5:1.000, -46:1.000 |
10:0.961, -41:0.993, 67:0.417 |
1:0.983, -50:0.968 |
1:0.468, -50:0.327 |
-1:0.920, -53:0.764 |
| chr6_qbl_hap6:2168808..2168927 | 19.257 | None | None (None) | None | None (None) | -15:1.000, 43:1.000 |
48:1.000 |
-10:0.919, 39:0.809 |
-8:0.302, 36:0.227 |
|
| chr19:58816586..58816705 | 19.231 | ZNF8|zinc finger protein 8 | -26269 (+) | ERVK3-1|endogenous retrovirus group K3, member 1 | 83 (+) | -27:1.000, 34:0.998 |
-22:0.998, 30:0.999, -65:0.740 |
-31:0.691 |
-31:0.999, 38:0.203 |
-34:0.223 |
| chr19:507880..507982 | 19.216 | TPGS1|tubulin polyglutamylase complex subunit 1 | -389 (+) | CDC34|cell division cycle 34 homolog (S. cerevisiae) | 23780 (+) | 25:1.000, -36:1.000 |
21:1.000, -31:1.000 |
30:0.980, -40:0.569 |
32:0.365 |
|
| chr5:57878758..57878880 | 19.215 | GAPT|GRB2-binding adaptor protein, transmembrane | -91525 (+) | RAB3C|RAB3C, member RAS oncogene family | 100 (+) | 22:0.996, -36:1.000 |
27:0.520 |
18:0.894, -31:0.980 |
15:0.499, -29:0.920 |
|
| chr19:36239188..36239320 | 19.213 | PSENEN|presenilin enhancer 2 homolog (C. elegans) | -2735 (+) | LIN37|lin-37 homolog (C. elegans) | 7 (+) | 44:1.000, -57:1.000 |
-18:0.995, 48:0.924, -53:0.997 |
39:0.920, -62:0.936 |
37:0.713, -64:0.239 |
|
| chr19:54618712..54618810 | 19.211 | NDUFA3|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa | -12595 (+) | PRPF31|PRP31 pre-mRNA processing factor 31 homolog (S. cerevisiae) | 66 (+) | 23:1.000, -28:0.996 |
-3:0.368, 18:0.978, -24:0.815 |
27:0.980 |
30:0.864 |
|
| chr9:102861336..102861475 | 19.196 | ERP44|endoplasmic reticulum protein 44 | -84 (-) | INVS|inversin | 76 (+) | 5:0.802, -53:1.000 |
-57:0.903 |
-48:0.968 |
-46:0.849 |
|
| chr22:20861775..20861881 | 19.194 | KLHL22|kelch-like 22 (Drosophila) | -11706 (-) | MED15|mediator complex subunit 15 | 38 (+) | 4:0.999, -24:0.998 |
-19:0.984 |
0:0.690, -28:0.449 |
0:0.215, -28:0.280 |
|
| chr17:73851561..73851679 | 19.180 | WBP2|WW domain binding protein 2 | -182 (-) | TRIM47|tripartite motif containing 47 | 22443 (-) | -30:1.000, 32:1.000 |
-26:1.000, 27:0.495 |
-35:0.983 |
-37:0.857, 39:0.857 |
|
| chr12:6961600..6961742 | 19.168 | USP5|ubiquitin specific peptidase 5 (isopeptidase T) | -342 (+) | TPI1|triosephosphate isomerase 1 | 14967 (+) | 17:0.998, -36:0.972, 88:0.540 |
12:0.750 |
21:0.450, 83:0.668 |
24:0.302, 81:0.849, -99:0.857 |
|
| chr6:116989934..116990028 | 19.147 | ZUFSP|zinc finger with UFM1-specific peptidase domain | -45 (-) | KPNA5|karyopherin alpha 5 (importin alpha 6) | 12386 (+) | 31:1.000 |
-9:0.787, 27:1.000 |
36:0.963, 69:0.205 |
38:0.754 |
|
| chr16:30908456..30908561 | 19.138 | CTF1|cardiotrophin 1 | -566 (+) | FBXL19-AS1|FBXL19 antisense RNA 1 (non-protein coding) | 26005 (-) | 10:0.989, -24:1.000 |
5:0.581, -20:0.954 |
14:0.691, -29:0.701 |
17:0.214, -31:0.849 |
|
| chr3:183892431..183892566 | 19.115 | DVL3|dishevelled, dsh homolog 3 (Drosophila) | -19133 (+) | AP2M1|adaptor-related protein complex 2, mu 1 subunit | 109 (+) | 15:0.675, 81:1.000 |
-67:0.964, 77:0.929 |
20:0.229, 86:0.919 |
22:0.863, 88:0.250 |
|
| chr3:4534896..4535021 | 19.114 | SUMF1|sulfatase modifying factor 1 | -25995 (-) | ITPR1|inositol 1,4,5-trisphosphate receptor, type 1 | 73 (+) | 25:0.998, -35:1.000 |
29:0.243, -40:1.000 |
20:0.302, -31:0.936 |
18:0.849, -28:0.917 |
|
| chr11:74109838..74109952 | 19.090 | PGM2L1|phosphoglucomutase 2-like 1 | -405 (-) | KCNE3|potassium voltage-gated channel, Isk-related family, member 3 | 68701 (-) | 28:0.999, -29:1.000 |
-33:0.979 |
-24:0.980, 24:0.868 |
21:0.431, -22:0.920 |
|
| chr22:29168586..29168693 | 19.086 | CHEK2|checkpoint kinase 2 | -30236 (-) | CCDC117|coiled-coil domain containing 117 | 5 (+) | -11:0.999, 30:0.992 |
-7:0.988, 34:0.808 |
-16:0.354 |
-18:0.368, -31:0.917 |
|
| chr2:54950519..54950644 | 19.084 | SPTBN1|spectrin, beta, non-erythrocytic 1 | -165016 (+) | EML6|echinoderm microtubule associated protein like 6 | 1567 (+) | 25:1.000, -32:0.999 |
30:0.997, -36:1.000 |
21:0.931, -27:0.506 |
21:0.233 |
18:0.702, -25:0.436 |
| chr20:62289872..62289989 | 19.057 | RTEL1|regulator of telomere elongation helicase 1 | -249 (+) | RTEL1|regulator of telomere elongation helicase 1 | 31218 (+) | -31:1.000, 31:1.000 |
-26:0.878, 27:0.998 |
-35:0.894, 36:0.716 |
-38:0.512, 38:0.512 |
|
| chr11:113644465..113644584 | 19.056 | ZW10|ZW10, kinetochore associated, homolog (Drosophila) | -40 (-) | CLDN25|claudin 25 | 5993 (+) | 16:1.000, -42:0.999 |
20:1.000, -47:0.998 |
11:0.980, -38:0.232 |
9:0.365 |
|
| chrX:106045753..106045869 | 19.040 | RNF128|ring finger protein 128 | -75803 (+) | TBC1D8B|TBC1 domain family, member 8B (with GRAM domain) | 170 (+) | -28:0.982, 31:0.999 |
-32:0.969 |
-23:0.309, 27:0.980 |
24:0.864 |
|
| chr18:56806960..56807081 | 19.034 | ZNF532|zinc finger protein 532 | -275671 (+) | SEC11C|SEC11 homolog C (S. cerevisiae) | 127 (+) | 28:0.999, -40:1.000 |
32:0.739, -45:1.000 |
23:0.885, -36:0.620 |
21:0.432 |
|
| chr19:51014491..51014633 | 19.030 | JOSD2|Josephin domain containing 2 | -152 (-) | ASPDH|aspartate dehydrogenase domain containing | 2593 (-) | -31:0.999, -62:0.961, 65:0.993 |
-26:0.974, 70:0.846 |
-35:0.868, 61:0.301, -66:0.894 |
-38:0.581, 58:0.365, -69:0.645 |
|
| chr12:861518..861634 | 19.007 | NINJ2|ninjurin 2 | -88861 (-) | WNK1|WNK lysine deficient protein kinase 1 | 580 (+) | -2:0.607, -43:1.000, 57:0.999 |
-6:0.932, -38:0.899, 62:0.995 |
-47:0.980 |
-50:0.864 |
|
| chr4:152682150..152682267 | 18.997 | PET112|PET112 homolog (yeast) | -67 (-) | FBXW7|F-box and WD repeat domain containing 7 | 591688 (-) | -14:0.999, 45:0.996 |
-19:0.998 |
-10:0.645, 40:0.270 |
-7:0.373 |
-7:0.238 |
| chr11:82997405..82997525 | 18.994 | CCDC90B|coiled-coil domain containing 90B | -60 (-) | DLG2|discs, large homolog 2 (Drosophila) | 395986 (-) | 33:1.000, -35:0.476, -70:0.405 |
-7:0.473, 29:0.590, -39:0.943 |
38:0.931, -65:0.657 |
37:0.923 |
40:0.702 |
| chr19:3435245..3435365 | 18.970 | NFIC|nuclear factor I/C (CCAAT-binding transcription factor) | -68690 (+) | 27534 (-) | 30:1.000, -33:0.987 |
26:1.000, -28:0.762 |
35:0.983 |
37:0.250, -40:0.427 |
||
| chr19:19887250..19887379 | 18.938 | LOC284440|uncharacterized LOC284440 | -109 (-) | ZNF506|zinc finger protein 506 | 45238 (-) | 11:1.000 |
16:0.998 |
7:0.980, -41:0.816, 96:0.816 |
4:0.857, -39:0.920, 98:0.917 |
|
| chr19:37861878..37862014 | 18.928 | HKR1|HKR1, GLI-Kruppel zinc finger family member | -36391 (+) | ZNF527|zinc finger protein 527 | 135 (+) | 9:0.952, -34:0.999 |
5:0.925, -38:0.999 |
-29:0.963 |
-27:0.855 |
|
| chr8:28747739..28747885 | 18.912 | INTS9|integrator complex subunit 9 | -115 (-) | HMBOX1|homeobox containing 1 | 105 (+) | -9:0.998, 62:0.997, -85:0.996 |
-14:0.808, 66:0.672, -90:0.580 |
-5:0.301, 57:0.980, -81:0.727 |
-2:0.920, 55:0.920, -78:0.702 |
|
| chr1:22110122..22110229 | 18.910 | USP48|ubiquitin specific peptidase 48 | -476 (-) | LDLRAD2|low density lipoprotein receptor class A domain containing 2 | 28631 (+) | -18:1.000, 25:1.000 |
-13:0.999, 21:0.998 |
-22:0.509, 30:0.919 |
-25:0.414, 32:0.857 |
|
| chr10:44144313..44144422 | 18.905 | ZNF32|zinc finger protein 32 | -61 (-) | CXCL12|chemokine (C-X-C motif) ligand 12 | 736132 (-) | -32:1.000, 41:1.000 |
-28:0.983, 36:0.620 |
-37:0.980, 45:0.968 |
-39:0.917, 48:0.855 |
|
| chrX:149736792..149736910 | 18.890 | MAMLD1|mastermind-like domain containing 1 | -123069 (+) | MTM1|myotubularin 1 | 195 (+) | -14:1.000 |
-18:0.983 |
-21:0.920 |
||
| chr2:241525722..241525833 | 18.885 | RNPEPL1|arginyl aminopeptidase (aminopeptidase B)-like 1 | -17669 (+) | CAPN10|calpain 10 | 452 (+) | -1:0.329, 17:1.000 |
13:0.996 |
22:0.894 |
24:0.499 |
|
| chr16:2205637..2205763 | 18.876 | RAB26|RAB26, member RAS oncogene family | -6955 (+) | TRAF7|TNF receptor-associated factor 7 | 81 (+) | -30:0.981, 38:1.000 |
42:0.457 |
33:0.774 |
-23:0.444, 31:0.713 |
|
| chr11:46134671..46134775 | 18.856 | GYLTL1B|glycosyltransferase-like 1B | -190507 (+) | PHF21A|PHD finger protein 21A | 8274 (-) | -26:1.000, 27:1.000 |
-22:1.000, 22:0.990 |
-31:0.716, 31:0.936 |
12:0.251 |
|
| chr20:48732474..48732588 | 18.853 | UBE2V1|ubiquitin-conjugating enzyme E2 variant 1 | -43 (-) | TMEM189|transmembrane protein 189 | 37659 (-) | -16:0.997, 18:1.000 |
14:1.000, -20:0.839 |
23:0.980 |
-9:0.857 |
|
| chr19:489011..489129 | 18.840 | ODF3L2|outer dense fiber of sperm tails 3-like 2 | -14142 (-) | MADCAM1|mucosal vascular addressin cell adhesion molecule 1 | 7406 (+) | 15:0.999, 56:0.656 |
11:0.995, 52:0.687 |
20:0.775 |
22:0.449 |
|
| chr8:74884639..74884773 | 18.839 | TCEB1|transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) | -195 (-) | TMEM70|transmembrane protein 70 | 3711 (+) | 6:1.000, 67:1.000 |
2:0.999, 63:0.998, -78:0.999 |
11:0.919, 72:0.936 |
13:0.864, 74:0.864 |
|
| chr8:145550578..145550697 | 18.838 | DGAT1|diacylglycerol O-acyltransferase 1 | -63 (-) | SCRT1|scratch homolog 1, zinc finger protein (Drosophila) | 9249 (-) | 9:1.000, -32:1.000 |
5:1.000, -36:1.000, 47:0.950 |
14:0.868, -27:0.868 |
||
| chr11:107436420..107436534 | 18.836 | ALKBH8|alkB, alkylation repair homolog 8 (E. coli) | -33 (-) | ELMOD1|ELMO/CED-12 domain containing 1 | 25334 (+) | -3:1.000 |
0:0.962, 31:0.217 |
-8:0.967 |
-10:0.215 |
|
| chr12:51477402..51477519 | 18.832 | CSRNP2|cysteine-serine-rich nuclear protein 2 | -84 (-) | TFCP2|transcription factor CP2 | 89107 (-) | -9:0.997, 33:1.000 |
-14:0.997, 28:0.967 |
-5:0.691, 37:0.604 |
||
| chr10:43048317..43048430 | 18.821 | ZNF37BP|zinc finger protein 37B, pseudogene | -171 (-) | ZNF33B|zinc finger protein 33B | 85625 (-) | -2:0.992, 43:1.000, -48:1.000 |
2:0.997, -43:0.997, 48:1.000 |
39:0.968, -52:0.967 |
39:0.901 |
36:0.754 |
| chr6_mcf_hap5:2257777..2257889 | 18.820 | None | None (None) | None | None (None) | -10:1.000, 48:1.000 |
53:1.000 |
-5:0.919, 44:0.809 |
-3:0.302, 41:0.227 |
|
| chr21:35288128..35288240 | 18.819 | ATP5O|ATP synthase, H+ transporting, mitochondrial F1 complex, O subunit | -92 (-) | MRPS6|mitochondrial ribosomal protein S6 SLC5A3|solute carrier family 5 (sodium/myo-inositol cotransporter), member 3 | 157657 (+) | -22:0.980, 31:0.992 |
-27:0.958 |
-18:0.296, 35:0.980 |
38:0.920 |
|
| chr19:46234175..46234306 | 18.802 | FBXO46|F-box protein 46 | -93 (-) | 37875 (-) | -4:1.000, -37:1.000, 74:0.847 |
0:0.951, -41:0.999, 79:0.325 |
-8:0.668, -32:0.607 |
-11:0.849 |
||
| chr3:113775373..113775496 | 18.788 | KIAA1407|KIAA1407 | -23 (-) | QTRTD1|queuine tRNA-ribosyltransferase domain containing 1 | 172 (+) | 17:0.992, -41:1.000 |
21:0.998, -46:0.965 |
-37:0.983 |
-34:0.920 |
|
| chr19:54606028..54606165 | 18.770 | OSCAR|osteoclast associated, immunoglobulin-like receptor | -1977 (-) | NDUFA3|NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 3, 9kDa | 70 (+) | 43:1.000, -52:1.000 |
47:1.000, -48:1.000 |
38:0.775, -57:0.775 |
36:0.436, -59:0.436 |
|
| chr8:67579541..67579690 | 18.744 | VCPIP1|valosin containing protein (p97)/p47 complex interacting protein 1 | -121 (-) | C8orf44-SGK3|C8orf44-SGK3 readthrough C8orf44|chromosome 8 open reading frame 44 | 207 (+) | -20:0.983, 77:0.999, -91:0.998 |
72:0.537, -96:0.923 |
81:0.229 |
-27:0.920, 84:0.863 |
|
| chr12:57881697..57881828 | 18.722 | ARHGAP9|Rho GTPase activating protein 9 | -8132 (-) | MARS|methionyl-tRNA synthetase | 30 (+) | -29:0.949, 31:1.000 |
-33:0.953, 36:1.000 |
27:0.691 |
24:0.223 |
|
| chr19:51611669..51611802 | 18.704 | CTU1|cytosolic thiouridylase subunit 1 homolog (S. pombe) | -94 (-) | SIGLEC9|sialic acid binding Ig-like lectin 9 | 16484 (+) | -16:0.985, -50:0.994, 51:0.986, -97:0.208 |
-11:0.996, -45:0.503, 56:0.741, 89:0.988 |
47:0.604, -54:0.619 |
||
| chr11:2421840..2421968 | 18.696 | CD81|CD81 molecule | -23357 (+) | TSSC4|tumor suppressing subtransferable candidate 4 | 92 (+) | -5:0.997, -44:0.999, 52:0.938 |
56:0.826 |
-1:0.894, -49:0.774 |
1:0.657, -51:0.702 |
|
| chr12:56710122..56710232 | 18.696 | CNPY2|canopy 2 homolog (zebrafish) | -198 (-) | PAN2|PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) | 17312 (-) | -29:1.000, 34:0.984 |
-24:1.000, 30:0.505 |
15:0.936, -33:0.963, 39:0.716 |
-33:0.280 |
12:0.260, -36:0.764, 41:0.512 |
| chr11:114310054..114310170 | 18.684 | RBM7|RNA binding motif protein 7 | -38710 (+) | REXO2|REX2, RNA exonuclease 2 homolog (S. cerevisiae) | 109 (+) | 27:0.991, -35:0.884 |
23:0.962, -30:0.796 |
|||
| chr17:27279373..27279515 | 18.674 | PHF12|PHD finger protein 12 | -948 (-) | SEZ6|seizure related 6 homolog (mouse) | 53581 (-) | 15:1.000, -74:0.988 |
11:0.785, -22:0.975, -69:0.860 |
20:0.907, -78:0.783 |
19:0.778 |
22:0.657, -81:0.348 |
| chr3:50388499..50388603 | 18.645 | NPRL2|nitrogen permease regulator-like 2 (S. cerevisiae) | -66 (-) | TMEM115|transmembrane protein 115 | 8359 (-) | 29:0.999, -32:0.998 |
-8:0.464, 24:0.481, -28:0.996 |
33:0.919, -37:0.334 |
36:0.857, -39:0.920 |
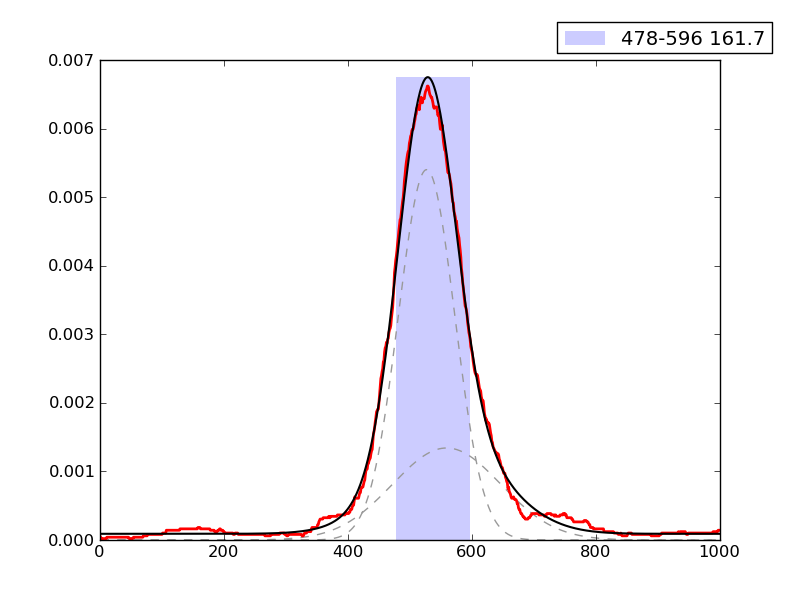
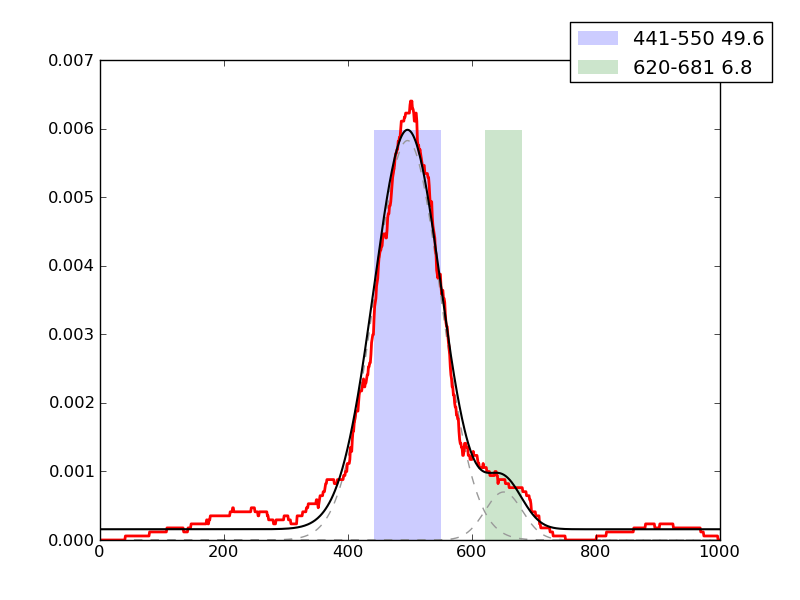
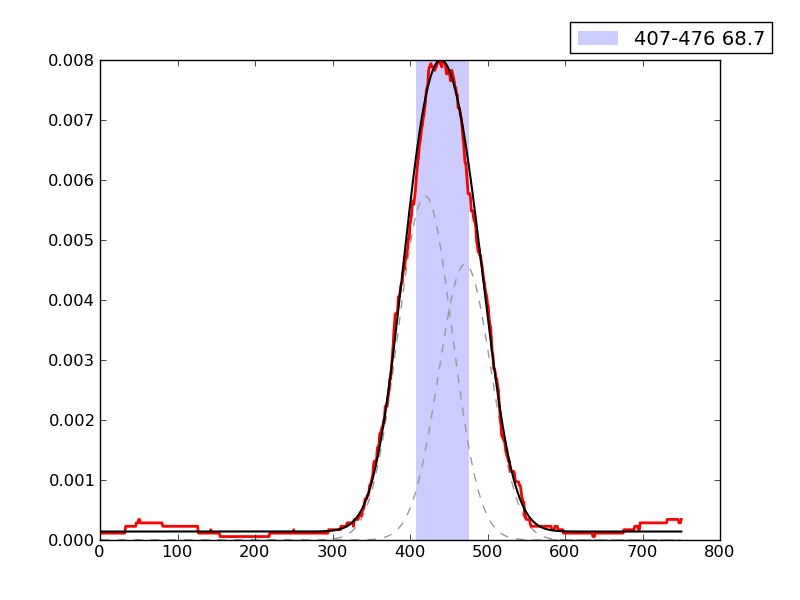
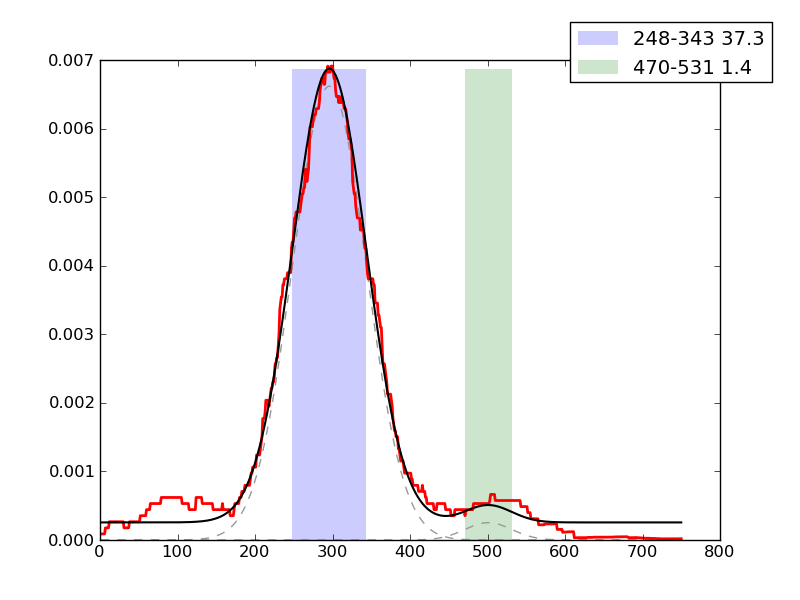
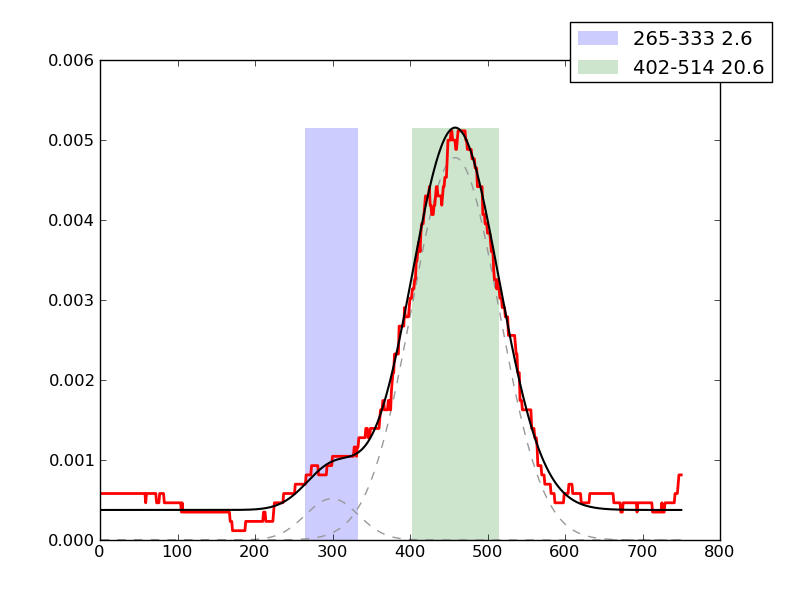
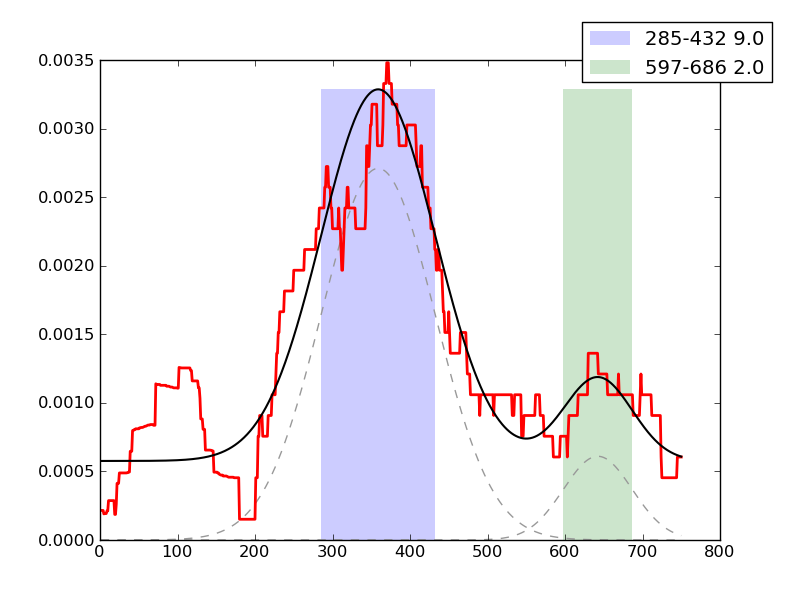
Examples of peaks fitted within regions
Complementary Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Motif Ensemble Enrichment (Motif Ensemble log-Likelihood Ratio) |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|---|
| denovo_WM_8 | 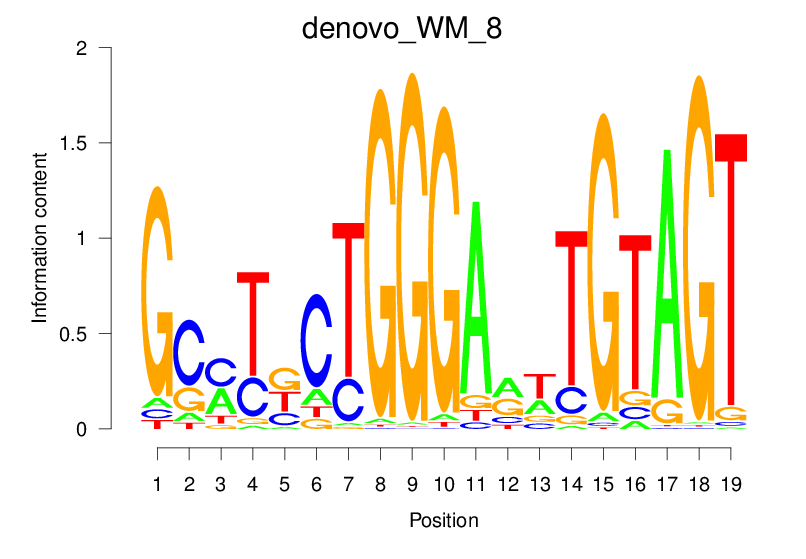 |
40.867 (1855.155) | 40.867 (1855.155) | 0.9180 | 0.6279 | 26.8263 | 3022/36955 |
| ZNF143.p2 | 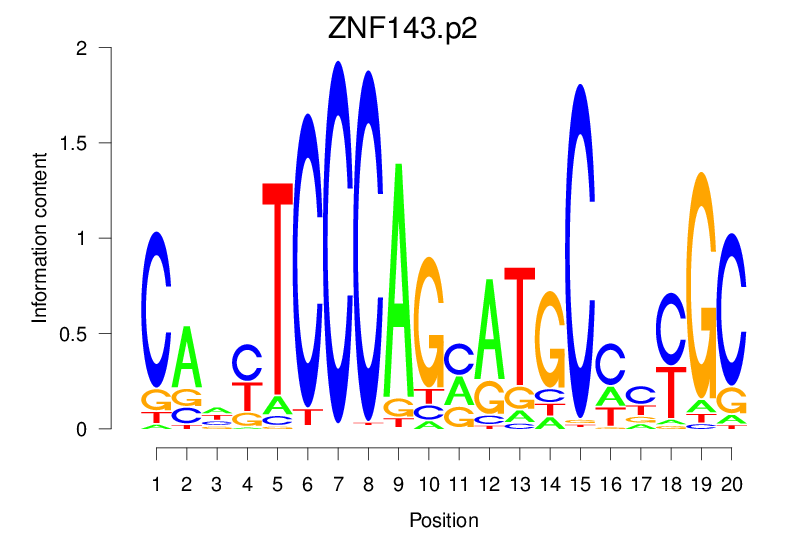 |
49.98 (1955.809) | 33.023 (1748.599) | 0.9402 | 0.5634 | 14.3887 | 5365/36955 |
| denovo_WM_12 | 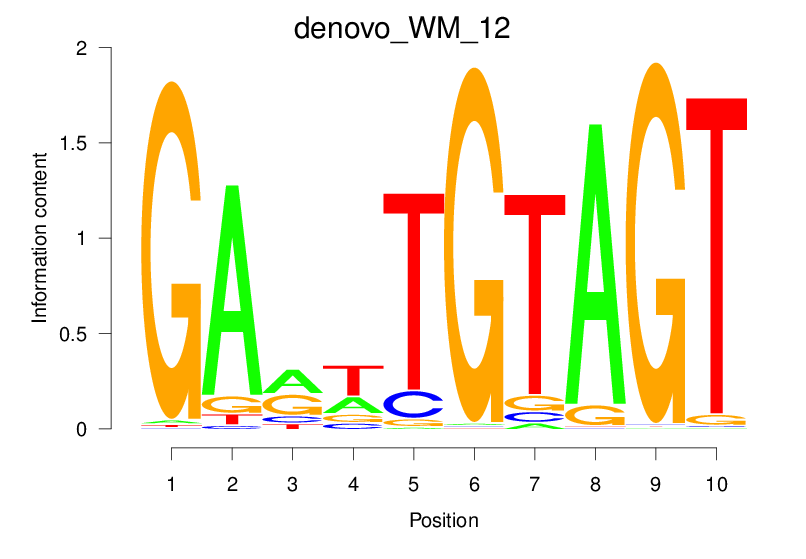 |
56.627 (2018.239) | 18.575 (1460.915) | 0.8717 | 0.5292 | 12.412 | 3761/36955 |
| STAT5{A,B}.p2 | 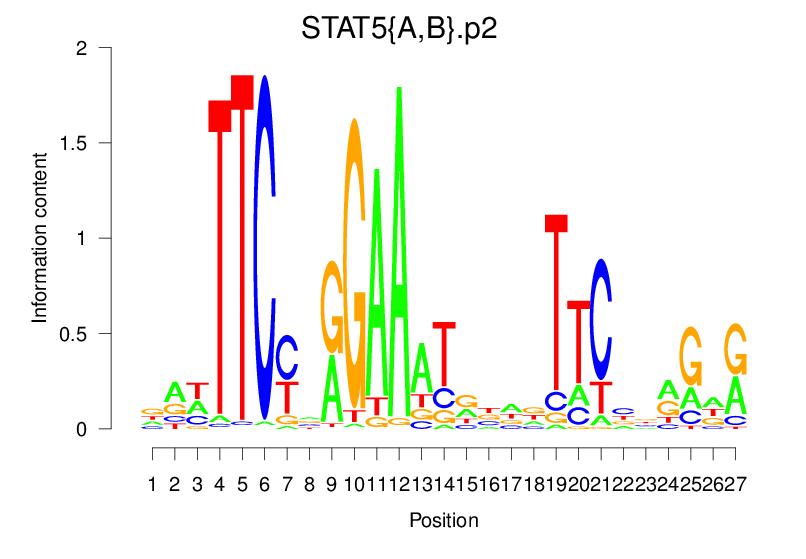 |
58.64 (2035.71) | 2.081 (366.381) | 0.5297 | 0.2175 | 8.2143 | 1039/36955 |
| denovo_WM_14 | 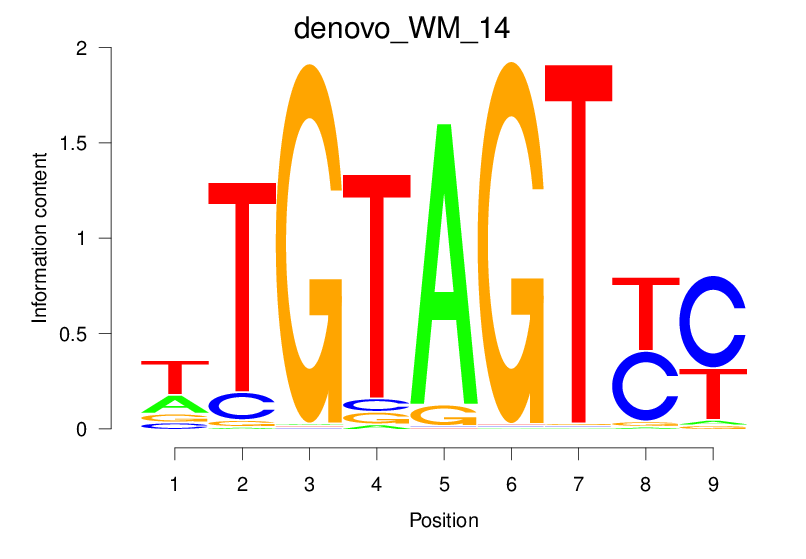 |
61.788 (2061.858) | 11.094 (1203.181) | 0.7994 | 0.4626 | 8.5302 | 4314/36955 |
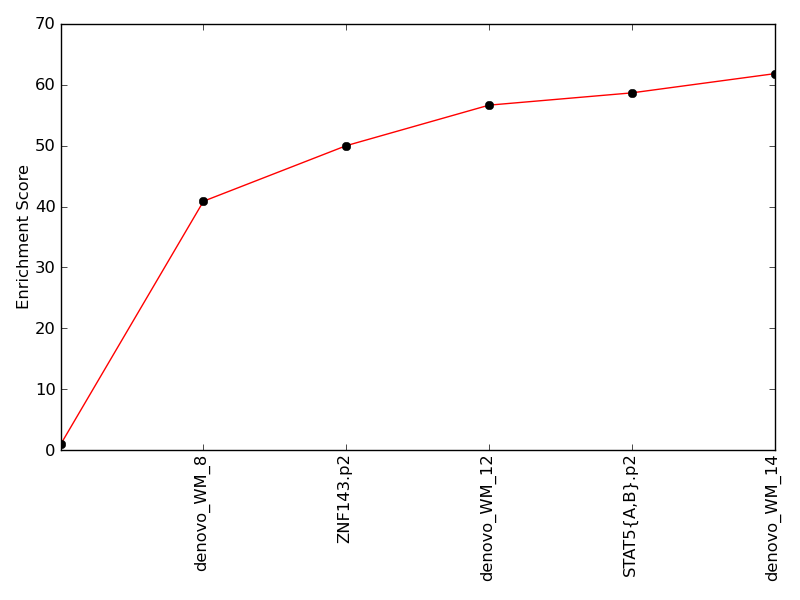
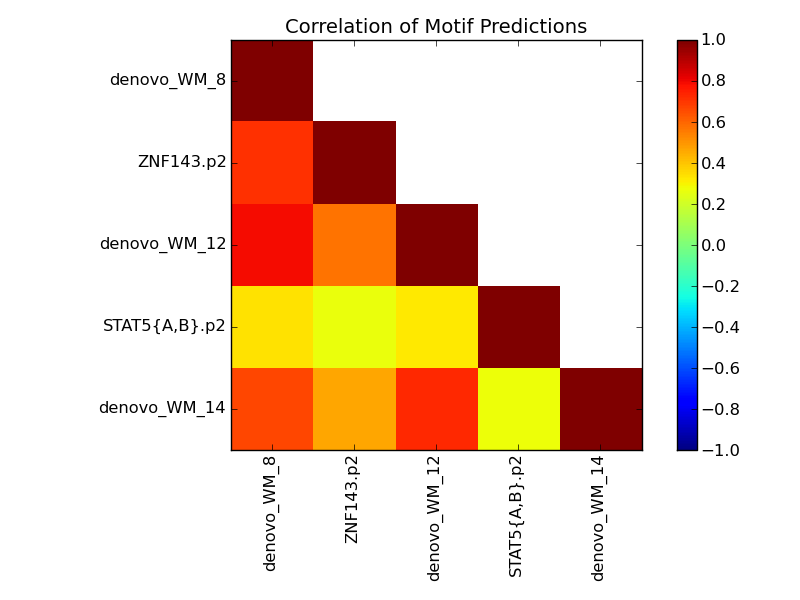
Contribution and Correlation Plots
Top Motifs Enriched In Peaks:
Motif Name |
Sequence Logo |
Enrichment (log-Likelihood Ratio) |
Precision and Recall |
Prediction - Observation Correlation |
Enrichment at Binding Sites |
Number of Positively Predicted Peaks |
|---|---|---|---|---|---|---|
| denovo_WM_8 |  |
40.867 (1855.155) | 0.9180 | 0.6279 | 26.8263 | 3022/36955 |
| denovo_WM_7 | 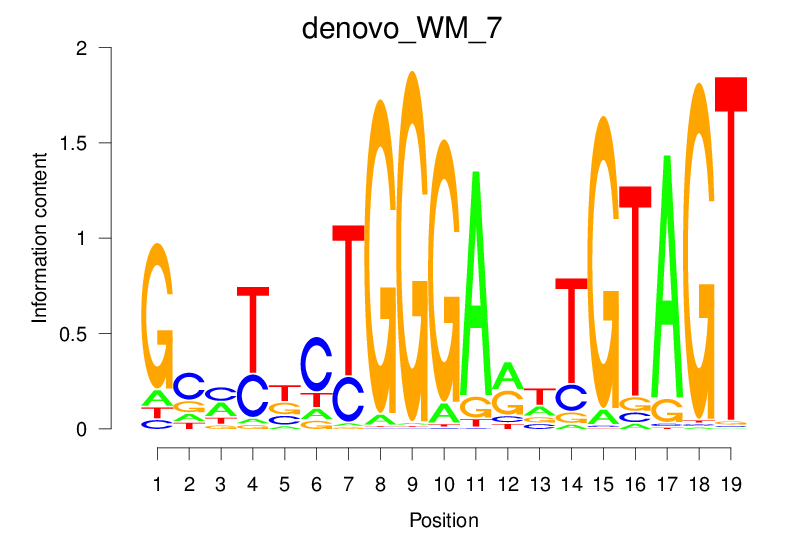 |
39.384 (1836.675) | 0.9117 | 0.6256 | 27.5404 | 2916/36955 |
| denovo_WM_18 | 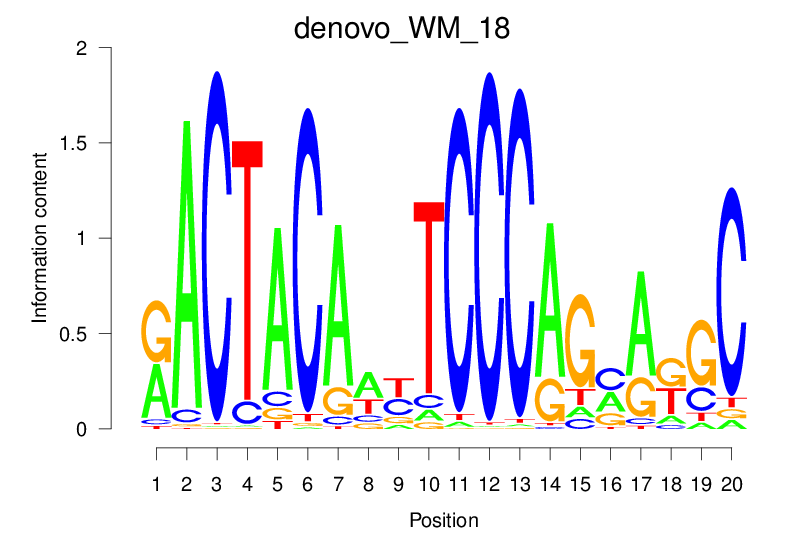 |
39.353 (1836.286) | 0.9124 | 0.632 | 29.5521 | 2780/36955 |
| denovo_WM_17 | 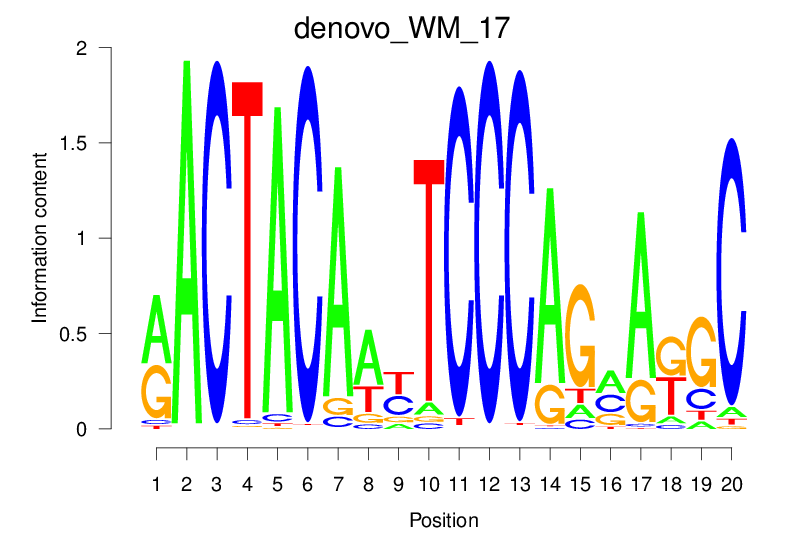 |
38.333 (1823.16) | 0.9021 | 0.64 | 42.3756 | 2215/36955 |
| denovo_WM_10 | 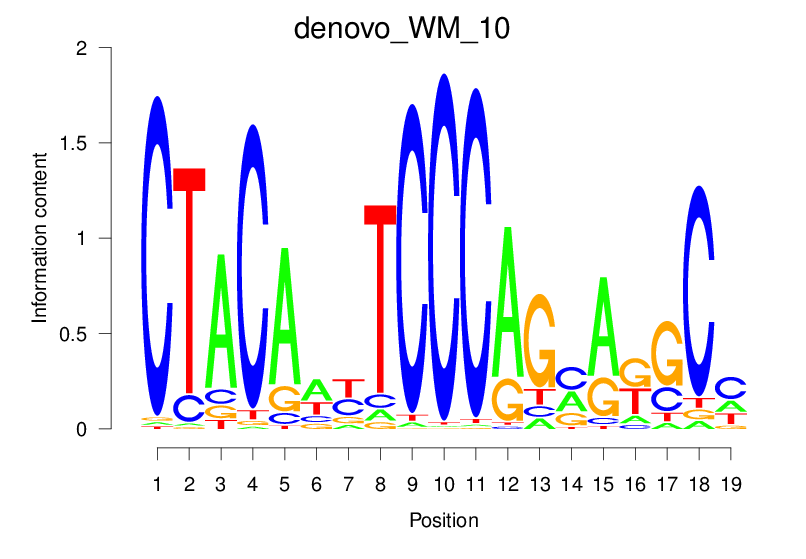 |
36.116 (1793.371) | 0.9289 | 0.6069 | 16.5148 | 4382/36955 |
| denovo_WM_16 | 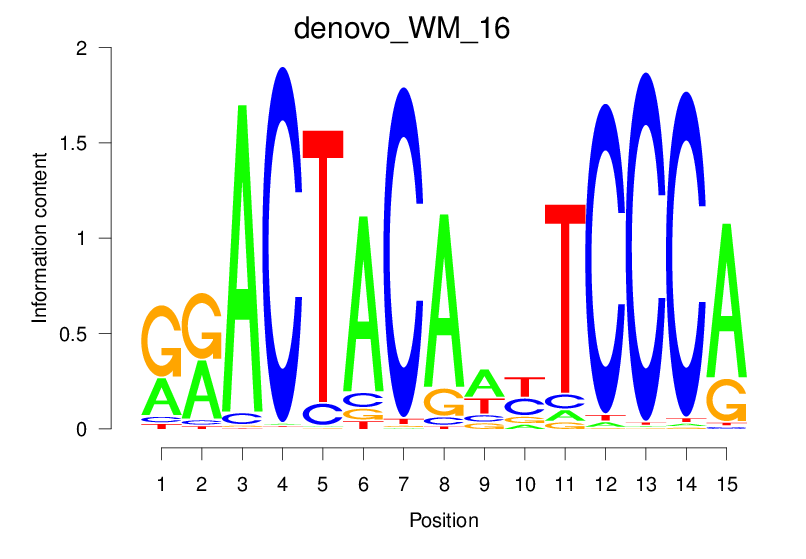 |
33.62 (1757.567) | 0.9036 | 0.6174 | 24.1159 | 2949/36955 |
| denovo_WM_15 | 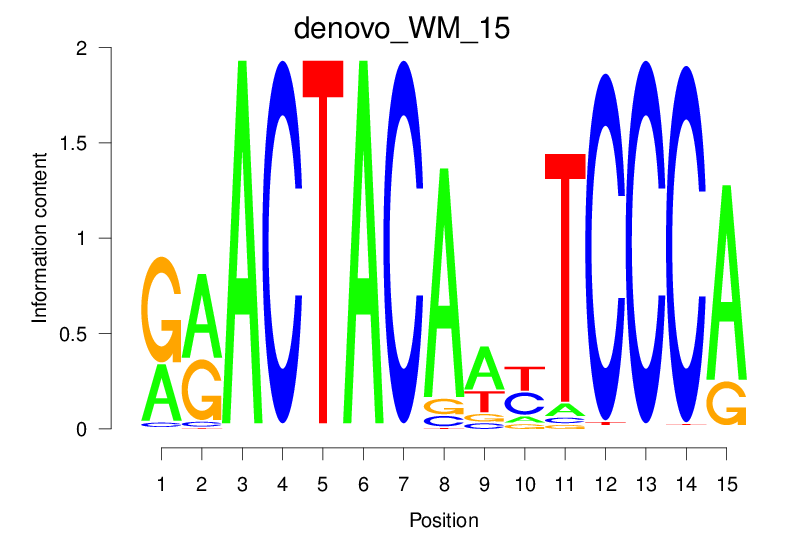 |
33.38 (1753.972) | 0.8943 | 0.6178 | 39.9949 | 2118/36955 |
| ZNF143.p2 |  |
33.023 (1748.599) | 0.9402 | 0.5634 | 14.3887 | 5365/36955 |
| denovo_WM_6 | 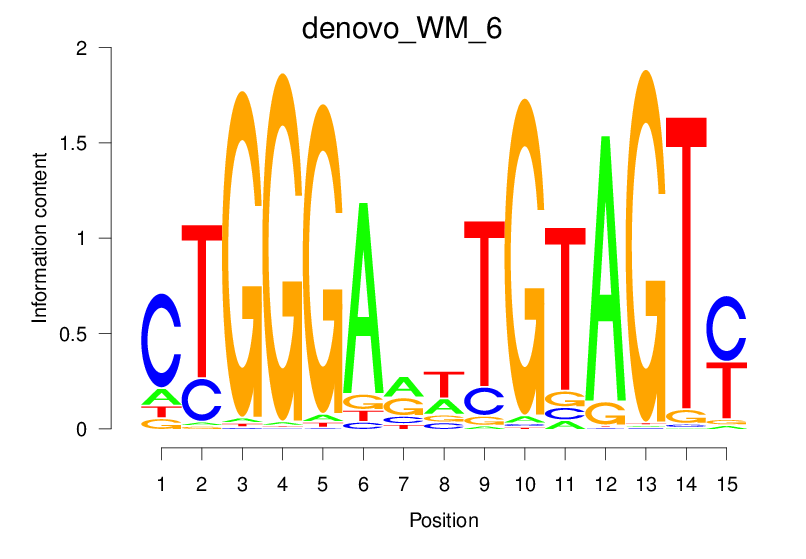 |
32.807 (1745.324) | 0.9061 | 0.6123 | 20.9514 | 3129/36955 |
| denovo_WM_5 | 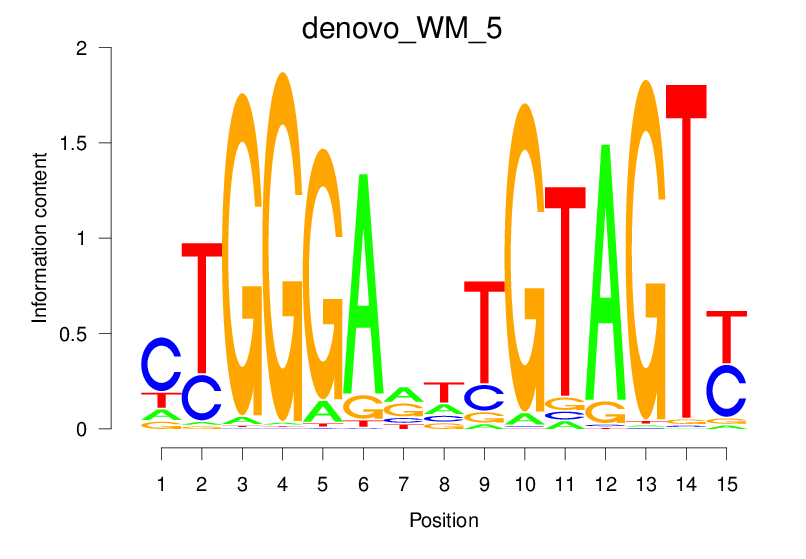 |
32.269 (1737.048) | 0.9067 | 0.6102 | 21.7348 | 3078/36955 |
Downloads
- Download Complete Analysis Report
- Download Peaks (with annotations and predicted binding sites)
- Download Detailed Report PDF about Peak Calling
- Download Detailed Report PDF about Motif Analysis