CRUNCH Results
Caution:
Crunch is not able to conduct motif analysis as not enough peaks (<200) were called to meaningfully analyse them. Supposedly low input data quality.
The significance level of Crunch can be lowered by increasing the false discovery rate (FDR) under the "Advanced options" menu.
The significance level of Crunch can be lowered by increasing the false discovery rate (FDR) under the "Advanced options" menu.
QC summary table
QC statistics for this dataset |
Comparison with ENCODE datasets |
||
Mapping |
|||
Fraction of mapped reads for the IP samples: |
0.717 | 39th percentile | |
Fraction of mapped reads for the background samples: |
0.866 | 74th percentile | |
ChIP enrichment signal |
|||
Noise level ChIP signal: |
0.262 | 75th percentile | |
Error in fit of enrichment distribution: |
1.922 | 97th percentile | |
Peak statistics |
|||
Number of peaks: |
533 | 7th percentile | |
Fraction of reads mapped to peaks: |
0.001 | 6th percentile | |
Quality Control, Mapping, Fragment Size Estimation:
ZZZ3 Replicate 1
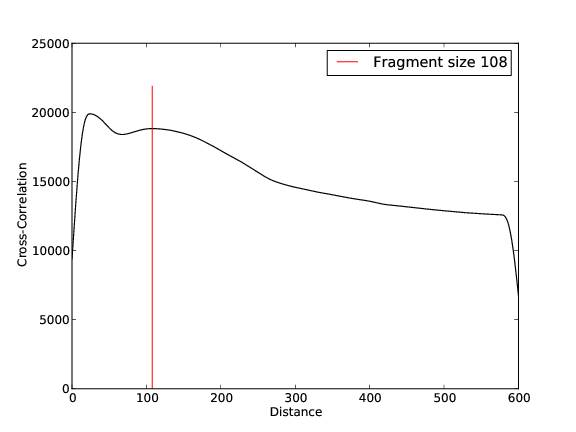
- 30,449,950 input reads (in FASTQ format).
- 24,887,763 reads after removal of low quality reads.
- 24,799,808 reads after removal of adapters and low complexity reads.
- 21,538,705 reads after mapping.
- Report PDF
ZZZ3 Replicate 2
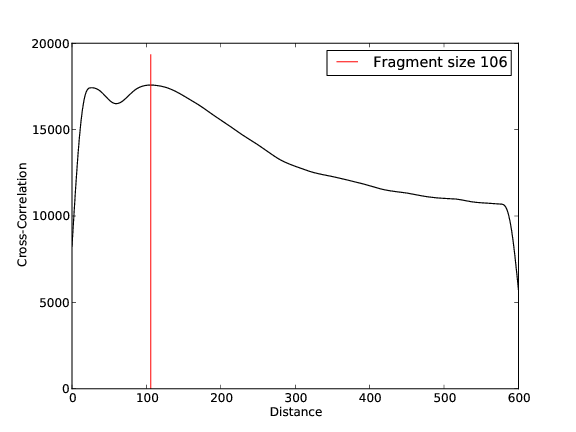
- 26,844,132 input reads (in FASTQ format).
- 22,325,260 reads after removal of low quality reads.
- 22,237,780 reads after removal of adapters and low complexity reads.
- 19,516,101 reads after mapping.
- Report PDF
BG Replicate 1
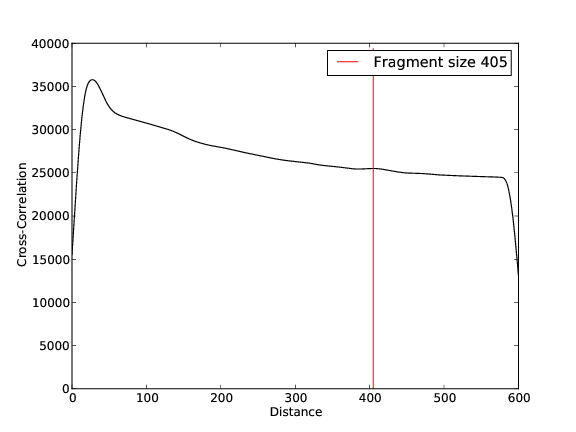
- 13,958,985 input reads (in FASTQ format).
- 13,816,004 reads after removal of low quality reads.
- 13,538,239 reads after removal of adapters and low complexity reads.
- 12,092,970 reads after mapping.
- Report PDF
Peak Calling:
- 1344 windows are significantly enriched at FDR=10% (z-value cut-off: 3.74)
- 1344 windows merged to 840 regions.
- 533 significant peaks fitted within 840 regions.
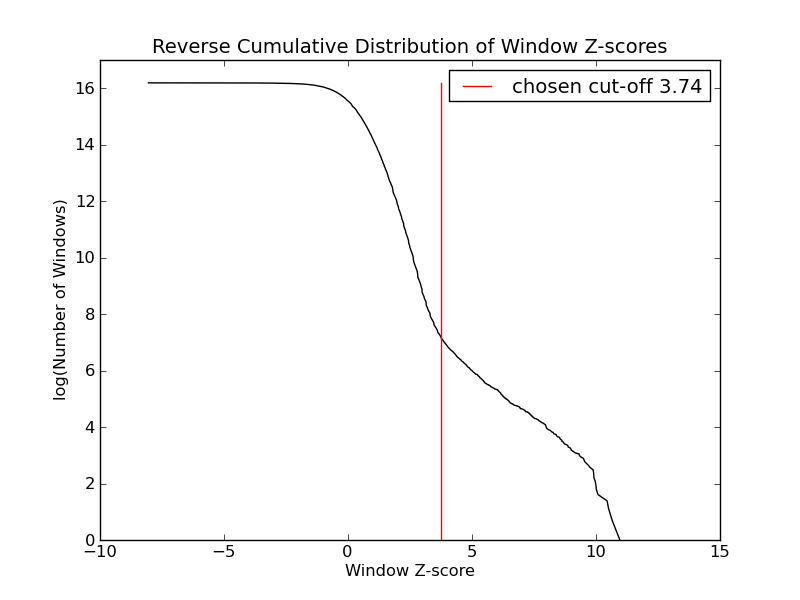
Top peaks:
| Coordinates | Z-score | Nearest Genes on the Left | Offset of Nearest TSS on the Left (Strand) | Nearest Genes on the Right | Offset of Nearest TSS on the Right (Strand) |
|---|---|---|---|---|---|
| chr3:138328322..138328493 | 10.509 | FAIM|Fas apoptotic inhibitory molecule | -489 (+) | PIK3CB|phosphoinositide-3-kinase, catalytic, beta polypeptide | 149777 (-) |
| chr3:57029598..57029742 | 10.454 | ARHGEF3|Rho guanine nucleotide exchange factor (GEF) 3 | -193759 (-) | SPATA12|spermatogenesis associated 12 | 64833 (+) |
| chr15:78994608..78994674 | 10.429 | CHRNB4|cholinergic receptor, nicotinic, beta 4 | -61068 (-) | MORF4L1|mortality factor 4 like 1 | 108186 (+) |
| chr1:155214561..155214709 | 9.946 | GBAP1|glucosidase, beta, acid pseudogene 1 GBA|glucosidase, beta, acid | -152 (-) | FAM189B|family with sequence similarity 189, member B | 10078 (-) |
| chr21:47321688..47321754 | 9.650 | PCBP3|poly(rC) binding protein 3 | -51833 (+) | COL6A1|collagen, type VI, alpha 1 | 79964 (+) |
| chr15:44580552..44580661 | 9.561 | FRMD5|FERM domain containing 5 | -93137 (-) | CASC4|cancer susceptibility candidate 4 | 293 (+) |
| chr10:110654324..110654460 | 9.497 | SORCS1|sortilin-related VPS10 domain containing receptor 1 | -1730016 (-) | XPNPEP1|X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | 1028817 (-) |
| chr1:97991269..97991390 | 9.426 | -730746 (+) | DPYD|dihydropyrimidine dehydrogenase | 395160 (-) | |
| chr21:34144231..34144396 | 9.253 | GCFC1|GC-rich sequence DNA-binding factor 1 | -239 (-) | C21orf49|chromosome 21 open reading frame 49 | 150 (+) |
| chr10:135063838..135063903 | 9.131 | VENTX|VENT homeobox | -12459 (+) | ADAM8|ADAM metallopeptidase domain 8 | 26495 (-) |
| chr1:207997326..207997464 | 9.050 | CD46|CD46 molecule, complement regulatory protein | -71912 (+) | 61928 (-) | |
| chr19:54729896..54729962 | 9.024 | LILRB3|leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | -2987 (-) | LILRA6|leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 6 LILRB3|leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 | 16732 (-) |
| chr8:71072844..71072987 | 8.996 | PRDM14|PR domain containing 14 | -89360 (-) | NCOA2|nuclear receptor coactivator 2 | 242632 (-) |
| chr15:44580748..44580844 | 8.966 | FRMD5|FERM domain containing 5 | -93327 (-) | CASC4|cancer susceptibility candidate 4 | 103 (+) |
| chr20:62362234..62362378 | 8.850 | ZGPAT|zinc finger, CCCH-type with G patch domain | -22910 (+) | LIME1|Lck interacting transmembrane adaptor 1 | 5676 (+) |
| chr11:107734925..107735069 | 8.808 | SLC35F2|solute carrier family 35, member F2 | -5084 (-) | RAB39A|RAB39A, member RAS oncogene family | 64238 (+) |
| chr19:597682..597862 | 8.775 | HCN2|hyperpolarization activated cyclic nucleotide-gated potassium channel 2 | -7859 (+) | POLRMT|polymerase (RNA) mitochondrial (DNA directed) | 35726 (-) |
| chr13:24845080..24845246 | 8.597 | SPATA13|spermatogenesis associated 13 | -265 (+) | C1QTNF9|C1q and tumor necrosis factor related protein 9 | 38554 (+) |
| chr19:597454..597625 | 8.527 | HCN2|hyperpolarization activated cyclic nucleotide-gated potassium channel 2 | -7626 (+) | POLRMT|polymerase (RNA) mitochondrial (DNA directed) | 35959 (-) |
| chr15:60691164..60691315 | 8.488 | ANXA2|annexin A2 | -1078 (-) | NARG2|NMDA receptor regulated 2 | 80035 (-) |
| chr10:91053390..91053532 | 8.390 | LIPA|lipase A, lysosomal acid, cholesterol esterase | -41839 (-) | IFIT2|interferon-induced protein with tetratricopeptide repeats 2 | 8294 (+) |
| chr18:11143598..11143777 | 8.389 | NAPG|N-ethylmaleimide-sensitive factor attachment protein, gamma | -617590 (+) | PIEZO2|piezo-type mechanosensitive ion channel component 2 | 5073 (-) |
| chr7:150452650..150452779 | 8.357 | LOC100128542|uncharacterized LOC100128542 | -5891 (+) | TMEM176B|transmembrane protein 176B | 44795 (-) |
| chr19:10538991..10539142 | 8.251 | PDE4A|phosphodiesterase 4A, cAMP-specific | -7836 (+) | PDE4A|phosphodiesterase 4A, cAMP-specific | 2361 (+) |
| chr7:74347543..74347662 | 8.156 | PMS2P5|postmeiotic segregation increased 2 pseudogene 5 | -40682 (+) | GATSL1|GATS protein-like 1 GATSL2|GATS protein-like 2 | 31478 (+) |
| chr6:37665826..37666002 | 8.149 | MDGA1|MAM domain containing glycosylphosphatidylinositol anchor 1 | -208 (-) | ZFAND3|zinc finger, AN1-type domain 3 | 121376 (+) |
| chr4:129474108..129474255 | 8.130 | PGRMC2|progesterone receptor membrane component 2 | -264198 (-) | PHF17|PHD finger protein 17 | 256689 (+) |
| chr1:161067356..161067510 | 8.039 | PVRL4|poliovirus receptor-related 4 | -8065 (-) | KLHDC9|kelch domain containing 9 | 737 (+) |
| chr7:72538077..72538201 | 8.028 | NSUN5P1|NOP2/Sun domain family, member 5 pseudogene 1 NSUN5P2|NOP2/Sun domain family, member 5 pseudogene 2 | -112897 (-) | GTF2IP1|general transcription factor IIi, pseudogene 1 | 30892 (+) |
| chr7:74684425..74684557 | 8.014 | GTF2IRD2B|GTF2I repeat domain containing 2B GTF2IRD2|GTF2I repeat domain containing 2 | -176117 (+) | GATSL2|GATS protein-like 2 GTF2IP1|general transcription factor IIi, pseudogene 1 | 183029 (-) |
| chr17:38601140..38601317 | 8.009 | IGFBP4|insulin-like growth factor binding protein 4 | -1409 (+) | TNS4|tensin 4 | 56618 (-) |
| chr8:82754716..82754831 | 7.915 | SNX16|sorting nexin 16 | -300 (-) | RALYL|RALY RNA binding protein-like | 2340686 (+) |
| chr11:9696736..9696916 | 7.908 | SWAP70|SWAP switching B-cell complex 70kDa subunit | -11120 (+) | LOC283104|uncharacterized LOC283104 | 83036 (+) |
| chr8:88673546..88673616 | 7.879 | CNBD1|cyclic nucleotide binding domain containing 1 | -794908 (+) | DCAF4L2|DDB1 and CUL4 associated factor 4-like 2 | 212678 (-) |
| chr4:155338318..155338477 | 7.827 | DCHS2|dachsous 2 (Drosophila) | -25949 (-) | DCHS2|dachsous 2 (Drosophila) | 74085 (-) |
| chr2:28407046..28407112 | 7.782 | LOC100302650|uncharacterized LOC100302650 | -293110 (-) | FOSL2|FOS-like antigen 2 | 208601 (+) |
| chr2:74517683..74517835 | 7.763 | MTHFD2|methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase | -92022 (+) | SLC4A5|solute carrier family 4, sodium bicarbonate cotransporter, member 5 | 24391 (-) |
| chr12:91996656..91996722 | 7.531 | DCN|decorin | -419884 (-) | BTG1|B-cell translocation gene 1, anti-proliferative | 542954 (-) |
| chr7:74898674..74898788 | 7.528 | GATSL2|GATS protein-like 2 GTF2IP1|general transcription factor IIi, pseudogene 1 | -31211 (-) | STAG3L1|stromal antigen 3-like 1 STAG3L2|stromal antigen 3-like 2 STAG3L3|stromal antigen 3-like 3 | 89839 (+) |
| chr12:46519051..46519175 | 7.498 | SCAF11|SR-related CTD-associated factor 11 | -133238 (-) | SLC38A1|solute carrier family 38, member 1 | 114500 (-) |
| chr18:58756119..58756258 | 7.467 | MC4R|melanocortin 4 receptor | -716188 (-) | CDH20|cadherin 20, type 2 | 401586 (+) |
| chr10:44503752..44503864 | 7.415 | ZNF32|zinc finger protein 32 | -359502 (-) | CXCL12|chemokine (C-X-C motif) ligand 12 | 376691 (-) |
| chr11:128563832..128563938 | 7.411 | FLI1|Friend leukemia virus integration 1 | -87 (+) | FLI1|Friend leukemia virus integration 1 | 76 (+) |
| chr18:29665272..29665403 | 7.373 | RNF125|ring finger protein 125 | -66531 (+) | RNF138|ring finger protein 138 | 6499 (+) |
| chr10:132158493..132158672 | 7.354 | GLRX3|glutaredoxin 3 | -223867 (+) | TCERG1L|transcription elongation regulator 1-like | 951238 (-) |
| chr1:180992158..180992313 | 7.184 | STX6|syntaxin 6 | -186 (-) | MR1|major histocompatibility complex, class I-related | 10325 (+) |
| chr10:132158791..132158910 | 7.152 | GLRX3|glutaredoxin 3 | -224135 (+) | TCERG1L|transcription elongation regulator 1-like | 950970 (-) |
| chr1:33921475..33921607 | 7.110 | PHC2|polyhomeotic homolog 2 (Drosophila) | -24933 (-) | ZSCAN20|zinc finger and SCAN domain containing 20 | 16710 (+) |
| chr3:114172958..114173118 | 7.098 | ZBTB20|zinc finger and BTB domain containing 20 | -70550 (-) | ZBTB20|zinc finger and BTB domain containing 20 | 170014 (-) |
| chr16:2088256..2088322 | 7.095 | SLC9A3R2|solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | -5009 (+) | NTHL1|nth endonuclease III-like 1 (E. coli) | 9553 (-) |
| chr9:23850701..23850838 | 7.062 | ELAVL2|ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B) | -24707 (-) | TUSC1|tumor suppressor candidate 1 | 1827564 (-) |
| chr7:30560435..30560609 | 7.052 | GGCT|gamma-glutamylcyclotransferase | -16121 (-) | GARS|glycyl-tRNA synthetase | 73658 (+) |
| chr5:75995555..75995730 | 6.995 | F2RL2|coagulation factor II (thrombin) receptor-like 2 | -76497 (-) | F2R|coagulation factor II (thrombin) receptor | 16314 (+) |
| chr19:55166445..55166585 | 6.984 | LILRB1|leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 1 | -24548 (+) | LILRB4|leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 4 | 7608 (+) |
| chrX:100353999..100354065 | 6.982 | CENPI|centromere protein I | -858 (+) | CENPI|centromere protein I | 765 (+) |
| chr11:64472039..64472180 | 6.978 | NRXN2|neurexin 2 | -61318 (-) | NRXN2|neurexin 2 | 18556 (-) |
| chr8:58136155..58136304 | 6.948 | IMPAD1|inositol monophosphatase domain containing 1 | -229803 (-) | FAM110B|family with sequence similarity 110, member B | 770928 (+) |
| chr1:111162628..111162779 | 6.947 | KCNA2|potassium voltage-gated channel, shaker-related subfamily, member 2 | -13721 (-) | KCNA2|potassium voltage-gated channel, shaker-related subfamily, member 2 | 11392 (-) |
| chr16:9133191..9133293 | 6.946 | USP7|ubiquitin specific peptidase 7 (herpes virus-associated) | -75947 (-) | C16orf72|chromosome 16 open reading frame 72 | 52298 (+) |
| chr12:2033942..2034121 | 6.939 | CACNA2D4|calcium channel, voltage-dependent, alpha 2/delta subunit 4 | -6130 (-) | DCP1B|DCP1 decapping enzyme homolog B (S. cerevisiae) | 79640 (-) |
| chr7:4745119..4745185 | 6.865 | FOXK1|forkhead box K1 | -22901 (+) | KIAA0415|KIAA0415 | 70109 (+) |
| chr2:124171685..124171770 | 6.863 | TSN|translin | -1658490 (+) | CNTNAP5|contactin associated protein-like 5 | 611137 (+) |
| chrX:70474879..70475058 | 6.861 | ZMYM3|zinc finger, MYM-type 3 | -482 (-) | ZMYM3|zinc finger, MYM-type 3 | 78 (-) |
| chr5:75995367..75995547 | 6.737 | F2RL2|coagulation factor II (thrombin) receptor-like 2 | -76312 (-) | F2R|coagulation factor II (thrombin) receptor | 16499 (+) |
| chr10:110654235..110654317 | 6.723 | SORCS1|sortilin-related VPS10 domain containing receptor 1 | -1729900 (-) | XPNPEP1|X-prolyl aminopeptidase (aminopeptidase P) 1, soluble | 1028933 (-) |
| chrX:129086741..129086876 | 6.688 | UTP14A|UTP14, U3 small nucleolar ribonucleoprotein, homolog A (yeast) | -46677 (+) | BCORL1|BCL6 corepressor-like 1 | 52398 (+) |
| chrX:61884345..61884443 | 6.670 | ZXDA|zinc finger, X-linked, duplicated A | -3947435 (-) | SPIN4|spindlin family, member 4 | 686805 (-) |
| chr2:43273929..43274078 | 6.668 | HAAO|3-hydroxyanthranilate 3,4-dioxygenase | -254252 (-) | ZFP36L2|zinc finger protein 36, C3H type-like 2 | 179342 (-) |
| chr8:81076200..81076338 | 6.644 | TPD52|tumor protein D52 | -83240 (-) | TPD52|tumor protein D52 | 7501 (-) |
| chr3:161750068..161750133 | 6.585 | OTOL1|otolin 1 | -535505 (+) | SI|sucrase-isomaltase (alpha-glucosidase) | 3046178 (-) |
| chr14:101083703..101083827 | 6.553 | BEGAIN|brain-enriched guanylate kinase-associated homolog (rat) | -47635 (-) | DLK1|delta-like 1 homolog (Drosophila) | 109458 (+) |
| chr21:34144093..34144192 | 6.538 | GCFC1|GC-rich sequence DNA-binding factor 1 | -68 (-) | C21orf49|chromosome 21 open reading frame 49 | 321 (+) |
| chr4:164165314..164165432 | 6.529 | NAF1|nuclear assembly factor 1 homolog (S. cerevisiae) | -77295 (-) | NPY1R|neuropeptide Y receptor Y1 | 88573 (-) |
| chr5:150439269..150439383 | 6.498 | GPX3|glutathione peroxidase 3 (plasma) | -39153 (+) | TNIP1|TNFAIP3 interacting protein 1 | 21248 (-) |
| chr3:105087014..105087160 | 6.479 | ALCAM|activated leukocyte cell adhesion molecule | -1265 (+) | CBLB|Cas-Br-M (murine) ecotropic retroviral transforming sequence b | 333967 (-) |
| chr17:28498919..28499080 | 6.462 | NSRP1|nuclear speckle splicing regulatory protein 1 | -55158 (+) | SLC6A4|solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | 63954 (-) |
| chr2:221996810..221996925 | 6.427 | SLC4A3|solute carrier family 4, anion exchanger, member 3 | -1504513 (+) | EPHA4|EPH receptor A4 | 440143 (-) |
| chr2:224822084..224822200 | 6.390 | WDFY1|WD repeat and FYVE domain containing 1 | -12077 (-) | MRPL44|mitochondrial ribosomal protein L44 | 11 (+) |
| chr4:124697596..124697738 | 6.313 | SPRY1|sprouty homolog 1, antagonist of FGF signaling (Drosophila) | -376994 (+) | ANKRD50|ankyrin repeat domain 50 | 936147 (-) |
| chr9:93862369..93862507 | 6.311 | SYK|spleen tyrosine kinase | -272705 (+) | AUH|AU RNA binding protein/enoyl-CoA hydratase | 261732 (-) |
| chr5:33443370..33443491 | 6.304 | TARS|threonyl-tRNA synthetase | -2291 (+) | ADAMTS12|ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 448615 (-) |
| chr17:57287291..57287400 | 6.294 | PRR11|proline rich 11 | -54225 (+) | SMG8|smg-8 homolog, nonsense mediated mRNA decay factor (C. elegans) | 52 (+) |
| chr4:79550404..79550584 | 6.287 | ANXA3|annexin A3 | -77586 (+) | BMP2K|BMP2 inducible kinase | 146989 (+) |
| chr4:160025606..160025753 | 6.252 | C4orf45|chromosome 4 open reading frame 45 | -69347 (-) | RAPGEF2|Rap guanine nucleotide exchange factor (GEF) 2 | 163318 (+) |
| chr15:30618460..30618639 | 6.229 | TJP1|tight junction protein 1 (zona occludens 1) | -503926 (-) | CHRFAM7A|CHRNA7 (cholinergic receptor, nicotinic, alpha 7, exons 5-10) and FAM7A (family with sequence similarity 7A, exons A-E) fusion | 67163 (-) |
| chr5:38832091..38832191 | 6.211 | LIFR|leukemia inhibitory factor receptor alpha | -236635 (-) | OSMR|oncostatin M receptor | 13818 (+) |
| chr11:55026376..55026476 | 6.210 | OR4C46|olfactory receptor, family 4, subfamily C, member 46 | -3511145 (+) | TRIM48|tripartite motif containing 48 | 3231 (+) |
| chr9:78790113..78790207 | 6.193 | PCSK5|proprotein convertase subtilisin/kexin type 5 | -284525 (+) | RFK|riboflavin kinase | 219231 (-) |
| chr6:20168726..20168791 | 6.143 | -330075 (+) | MBOAT1|membrane bound O-acyltransferase domain containing 1 | 43914 (-) | |
| chr11:128284395..128284568 | 6.140 | KIRREL3-AS3|KIRREL3 antisense RNA 3 (non-protein coding) | -1411176 (+) | ETS1|v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) | 107482 (-) |
| chr2:3622716..3622825 | 6.048 | RNASEH1|ribonuclease H1 | -16828 (-) | 9 (-) | |
| chr3:164914670..164914777 | 6.034 | SLITRK3|SLIT and NTRK-like family, member 3 | -218 (-) | BCHE|butyrylcholinesterase | 640470 (-) |
| chr3:154147943..154148025 | 6.011 | GPR149|G protein-coupled receptor 149 | -481 (-) | MME|membrane metallo-endopeptidase | 649465 (+) |
| chr3:114173758..114173902 | 5.989 | ZBTB20|zinc finger and BTB domain containing 20 | -71342 (-) | ZBTB20|zinc finger and BTB domain containing 20 | 169222 (-) |
| chrX:80065131..80065255 | 5.960 | FAM46D|family with sequence similarity 46, member D | -389231 (+) | BRWD3|bromodomain and WD repeat domain containing 3 | 14 (-) |
| chr14:68623117..68623261 | 5.929 | RAD51B|RAD51 homolog B (S. cerevisiae) | -336669 (+) | ZFP36L1|zinc finger protein 36, C3H type-like 1 | 636531 (-) |
| chr5:110867565..110867641 | 5.899 | STARD4|StAR-related lipid transfer (START) domain containing 4 | -19309 (-) | C5orf13|chromosome 5 open reading frame 13 | 224344 (-) |
| chr7:83824287..83824440 | 5.897 | SEMA3A|sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | -41 (-) | SEMA3D|sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D | 926883 (-) |
| chr3:43275122..43275256 | 5.865 | C3orf39|chromosome 3 open reading frame 39 | -127638 (-) | SNRK|SNF related kinase | 52880 (+) |
| chr4:1874226..1874386 | 5.847 | WHSC1|Wolf-Hirschhorn syndrome candidate 1 | -1108 (+) | WHSC1|Wolf-Hirschhorn syndrome candidate 1 | 20202 (+) |
| chr11:128761315..128761420 | 5.800 | KCNJ5|potassium inwardly-rectifying channel, subfamily J, member 5 | -27 (+) | C11orf45|chromosome 11 open reading frame 45 | 13162 (-) |
| chr6:469500..469572 | 5.784 | IRF4|interferon regulatory factor 4 | -77772 (+) | HUS1B|HUS1 checkpoint homolog b (S. pombe) | 187417 (-) |
| chr1:247526420..247526512 | 5.745 | ZNF496|zinc finger protein 496 | -31449 (-) | NLRP3|NLR family, pyrin domain containing 3 | 53003 (+) |
| chr22:44695917..44696000 | 5.742 | PARVG|parvin, gamma | -118702 (+) | KIAA1644|KIAA1644 | 12772 (-) |
| chr2:136788709..136788836 | 5.687 | DARS|aspartyl-tRNA synthetase | -45556 (-) | CXCR4|chemokine (C-X-C motif) receptor 4 | 85026 (-) |
| chr11:51288027..51288112 | 5.686 | OR4C12|olfactory receptor, family 4, subfamily C, member 12 | -1283999 (-) | OR4A5|olfactory receptor, family 4, subfamily A, member 5 | 124378 (-) |
| chr1:44679012..44679135 | 5.679 | KLF17|Kruppel-like factor 17 | -94524 (+) | DMAP1|DNA methyltransferase 1 associated protein 1 | 70 (+) |
| chr12:72092362..72092491 | 5.674 | TMEM19|transmembrane protein 19 | -12199 (+) | RAB21|RAB21, member RAS oncogene family | 56229 (+) |
| chr1:37908312..37908379 | 5.672 | GRIK3|glutamate receptor, ionotropic, kainate 3 | -408570 (-) | ZC3H12A|zinc finger CCCH-type containing 12A | 31752 (+) |
| chr16:46725789..46725939 | 5.650 | ORC6|origin recognition complex, subunit 6 PPA2|pyrophosphatase (inorganic) 2 | -2267 (+) | MYLK3|myosin light chain kinase 3 | 56298 (-) |
| chr1:193909161..193909228 | 5.623 | B3GALT2|UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 2 | -753462 (-) | KCNT2|potassium channel, subfamily T, member 2 | 2668331 (-) |
| chr7:29235158..29235302 | 5.617 | CHN2|chimerin (chimaerin) 2 | -789 (+) | CHN2|chimerin (chimaerin) 2 | 284255 (+) |
| chr8:47368605..47368677 | 5.614 | POTEA|POTE ankyrin domain family, member A | -4221036 (+) | KIAA0146|KIAA0146 | 804854 (+) |
| chr6:18749809..18749943 | 5.613 | RNF144B|ring finger protein 144B | -362209 (+) | ID4|inhibitor of DNA binding 4, dominant negative helix-loop-helix protein | 1087759 (+) |
| chr2:198920007..198920110 | 5.587 | PLCL1|phospholipase C-like 1 | -250688 (+) | SATB2|SATB homeobox 2 | 1293792 (-) |
| chr1:226899052..226899176 | 5.566 | C1orf95|chromosome 1 open reading frame 95 | -162497 (+) | ITPKB|inositol-trisphosphate 3-kinase B | 27824 (-) |
| chr16:35215926..35216010 | 5.557 | LOC653550|TP53-target gene 3 protein-like TP53TG3B|TP53 target 3B TP53TG3|TP53 target 3 | -1953835 (+) | SHCBP1|SHC SH2-domain binding protein 1 | 11439304 (-) |
| chr3:143695332..143695476 | 5.550 | C3orf58|chromosome 3 open reading frame 58 | -3187 (+) | PLOD2|procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 2183403 (-) |
| chr8:38741287..38741403 | 5.521 | TACC1|transforming, acidic coiled-coil containing protein 1 | -96577 (+) | PLEKHA2|pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 2 | 17466 (+) |
| chr8:67434005..67434161 | 5.521 | C8orf46|chromosome 8 open reading frame 46 | -28593 (+) | LOC645895|uncharacterized LOC645895 MYBL1|v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | 91391 (-) |
| chr19:28047770..28047869 | 5.502 | ZNF254|zinc finger protein 254 | -3777850 (+) | 236569 (+) | |
| chr7:62021778..62021880 | 5.489 | ZNF716|zinc finger protein 716 | -4511947 (+) | ZNF727|zinc finger protein 727 | 1483991 (+) |
| chrX:62005975..62006052 | 5.465 | ZXDA|zinc finger, X-linked, duplicated A | -4069054 (-) | SPIN4|spindlin family, member 4 | 565186 (-) |
| chr15:40169058..40169140 | 5.448 | -93993 (+) | 43186 (-) | ||
| chr1:89487401..89487580 | 5.439 | CCBL2|cysteine conjugate-beta lyase 2 RBMXL1|RNA binding motif protein, X-linked-like 1 | -28848 (-) | GBP3|guanylate binding protein 3 | 990 (-) |
| chr9:70076696..70076800 | 5.434 | ANKRD20A1|ankyrin repeat domain 20 family, member A1 ANKRD20A4|ankyrin repeat domain 20 family, member A4 | -694768 (+) | FOXD4L5|forkhead box D4-like 5 | 102066 (-) |
| chr9:70327437..70327540 | 5.433 | FOXD4L5|forkhead box D4-like 5 | -148674 (-) | FOXD4L2|forkhead box D4-like 2 FOXD4L4|forkhead box D4-like 4 | 102242 (-) |
| chr22:27044843..27044955 | 5.425 | CRYBA4|crystallin, beta A4 | -26968 (+) | MIAT|myocardial infarction associated transcript (non-protein coding) | 8573 (+) |
| chr3:90447914..90448014 | 5.409 | EPHA3|EPH receptor A3 | -1291114 (+) | PROS1|protein S (alpha) | 3244712 (-) |
| chr9:21990558..21990700 | 5.400 | CDKN2A|cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | -15498 (-) | CDKN2A|cyclin-dependent kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 3720 (-) |
| chr10:18647399..18647526 | 5.395 | CACNB2|calcium channel, voltage-dependent, beta 2 subunit | -17849 (+) | CACNB2|calcium channel, voltage-dependent, beta 2 subunit | 42101 (+) |
| chrX:951422..951597 | 5.367 | SHOX|short stature homeobox | -366431 (+) | CRLF2|cytokine receptor-like factor 2 | 380075 (-) |
| chrX:139844335..139844458 | 5.356 | RP1-177G6.2|uncharacterized LOC286411 | -52469 (+) | CDR1|cerebellar degeneration-related protein 1, 34kDa | 22256 (-) |
| chr3:90326980..90327084 | 5.355 | EPHA3|EPH receptor A3 | -1170182 (+) | PROS1|protein S (alpha) | 3365644 (-) |
| chr9:42819082..42819187 | 5.354 | FOXD4L2|forkhead box D4-like 2 FOXD4L4|forkhead box D4-like 4 | -101901 (+) | ANKRD20A1|ankyrin repeat domain 20 family, member A1 ANKRD20A2|ankyrin repeat domain 20 family, member A2 ANKRD20A3|ankyrin repeat domain 20 family, member A3 | 314353 (-) |
| chr2:65529186..65529348 | 5.353 | ACTR2|ARP2 actin-related protein 2 homolog (yeast) | -74332 (+) | SPRED2|sprouty-related, EVH1 domain containing 2 | 64506 (-) |
| chr2:181350207..181350273 | 5.347 | CWC22|CWC22 spliceosome-associated protein homolog (S. cerevisiae) | -478426 (-) | UBE2E3|ubiquitin-conjugating enzyme E2E 3 | 494985 (+) |
| chrX:9979631..9979800 | 5.333 | LOC100288814|uncharacterized LOC100288814 | -44318 (+) | WWC3|WWC family member 3 | 4078 (+) |
| chr21:25485841..25485944 | 5.332 | NCAM2|neural cell adhesion molecule 2 | -3115119 (+) | FLJ42200|FLJ42200 protein | 315176 (+) |
| chr7:141431543..141431654 | 5.328 | WEE2|WEE1 homolog 2 (S. pombe) | -23446 (+) | SSBP1|single-stranded DNA binding protein 1 | 6666 (+) |
| chrX:61827420..61827519 | 5.322 | ZXDA|zinc finger, X-linked, duplicated A | -3890510 (-) | SPIN4|spindlin family, member 4 | 743730 (-) |
| chrY:13680931..13681032 | 5.318 | TSPY1|testis specific protein, Y-linked 1 TSPY3|testis specific protein, Y-linked 3 | -4315445 (+) | TTTY15|testis-specific transcript, Y-linked 15 (non-protein coding) | 1093268 (+) |
| chr8:22668376..22668556 | 5.307 | EGR3|early growth response 3 | -117682 (-) | PEBP4|phosphatidylethanolamine-binding protein 4 | 116858 (-) |
| chr11:48845502..48845605 | 5.306 | OR4A47|olfactory receptor, family 4, subfamily A, member 47 | -335247 (+) | LOC120824|tripartite motif-containing protein ENSP00000309378-like | 157693 (-) |
| chrX:40653382..40653467 | 5.302 | MED14|mediator complex subunit 14 | -58295 (-) | USP9X|ubiquitin specific peptidase 9, X-linked | 291448 (+) |
| chr16:51474946..51475114 | 5.292 | SALL1|sal-like 1 (Drosophila) | -289618 (-) | LOC388276|hCG2045437 | 632816 (-) |
| chr22:50338979..50339093 | 5.286 | CRELD2|cysteine-rich with EGF-like domains 2 | -26661 (+) | PIM3|pim-3 oncogene | 15113 (+) |
| chr1:199498642..199498749 | 5.285 | LOC100131234|familial acute myelogenous leukemia related factor | -592148 (-) | NR5A2|nuclear receptor subfamily 5, group A, member 2 | 498174 (+) |
| chr3:49827476..49827607 | 5.247 | IP6K1|inositol hexakisphosphate kinase 1 | -3567 (-) | CDHR4|cadherin-related family member 4 | 9719 (-) |
| chr7:44885631..44885756 | 5.223 | PPIA|peptidylprolyl isomerase A (cyclophilin A) | -49377 (+) | H2AFV|H2A histone family, member V | 1885 (-) |
| chr1:161706255..161706394 | 5.221 | FCRLB|Fc receptor-like B | -13869 (+) | DUSP12|dual specificity phosphatase 12 | 13242 (+) |
| chr11:11405322..11405403 | 5.218 | LOC729013|uncharacterized LOC729013 | -525576 (+) | GALNTL4|UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase-like 4 | 238216 (-) |
| chr17:55776466..55776620 | 5.212 | MSI2|musashi homolog 2 (Drosophila) | -442170 (+) | MRPS23|mitochondrial ribosomal protein S23 | 150861 (-) |
| chr8:73330593..73330754 | 5.210 | TRPA1|transient receptor potential cation channel, subfamily A, member 1 | -342839 (-) | KCNB2|potassium voltage-gated channel, Shab-related subfamily, member 2 | 118970 (+) |
| chr2:85960015..85960124 | 5.201 | GNLY|granulysin | -38589 (+) | ATOH8|atonal homolog 8 (Drosophila) | 20936 (+) |
| chr1:161067540..161067634 | 5.200 | PVRL4|poliovirus receptor-related 4 | -8219 (-) | KLHDC9|kelch domain containing 9 | 583 (+) |
| chr5:159953827..159954006 | 5.200 | PTTG1|pituitary tumor-transforming 1 | -105013 (+) | ATP10B|ATPase, class V, type 10B | 158423 (-) |
| chr14:19044556..19044656 | 5.199 | None (None) | OR11H12|olfactory receptor, family 11, subfamily H, member 12 | 332951 (+) | |
| chr11:9696983..9697068 | 5.197 | SWAP70|SWAP switching B-cell complex 70kDa subunit | -11319 (+) | LOC283104|uncharacterized LOC283104 | 82837 (+) |
| chrY:901434..901601 | 5.193 | SHOX|short stature homeobox | -366439 (+) | CRLF2|cytokine receptor-like factor 2 | 380067 (-) |
| chr5:125754932..125755070 | 5.189 | GRAMD3|GRAM domain containing 3 | -59187 (+) | GRAMD3|GRAM domain containing 3 | 3842 (+) |
| chr5:46347132..46347256 | 5.181 | HCN1|hyperpolarization activated cyclic nucleotide-gated potassium channel 1 | -650975 (-) | EMB|embigin | 3390005 (-) |
| chr19:51003190..51003344 | 5.167 | FAM71E1|family with sequence similarity 71, member E1 | -23265 (-) | JOSD2|Josephin domain containing 2 | 11143 (-) |
| chrX:61806698..61806808 | 5.163 | ZXDA|zinc finger, X-linked, duplicated A | -3869794 (-) | SPIN4|spindlin family, member 4 | 764446 (-) |
| chr8:74092644..74092718 | 5.140 | C8orf84|chromosome 8 open reading frame 84 | -87239 (-) | LOC100130301|uncharacterized LOC100130301 | 79055 (-) |
| chr19_gl000208_random:132..278 | 5.117 | None | None (None) | None | None (None) |
| chr8:46958086..46958159 | 5.115 | POTEA|POTE ankyrin domain family, member A | -3810517 (+) | KIAA0146|KIAA0146 | 1215373 (+) |
| chrX:119443496..119443627 | 5.092 | -58871 (+) | FAM70A|family with sequence similarity 70, member A | 1724 (-) | |
| chr16:34002116..34002232 | 5.089 | LOC653550|TP53-target gene 3 protein-like TP53TG3B|TP53 target 3B TP53TG3|TP53 target 3 | -740041 (+) | SHCBP1|SHC SH2-domain binding protein 1 | 12653098 (-) |
| chr8:47368539..47368604 | 5.088 | POTEA|POTE ankyrin domain family, member A | -4220966 (+) | KIAA0146|KIAA0146 | 804924 (+) |
| chr9:109626307..109626417 | 5.084 | ZNF462|zinc finger protein 462 | -998 (+) | ZNF462|zinc finger protein 462 | 60107 (+) |
| chr9:100925968..100926119 | 5.078 | TRIM14|tripartite motif containing 14 | -44499 (-) | CORO2A|coronin, actin binding protein, 2A | 9130 (-) |
| chr4:146859998..146860111 | 5.062 | -86 (-) | LSM6|LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) | 236813 (+) | |
| chrY:13704338..13704413 | 5.056 | TSPY1|testis specific protein, Y-linked 1 TSPY3|testis specific protein, Y-linked 3 | -4338839 (+) | TTTY15|testis-specific transcript, Y-linked 15 (non-protein coding) | 1069874 (+) |
| chr21:47065907..47066036 | 5.039 | PCBP3|poly(rC) binding protein 3 | -2342 (+) | PCBP3|poly(rC) binding protein 3 | 203917 (+) |
| chr3:57029815..57029995 | 5.035 | ARHGEF3|Rho guanine nucleotide exchange factor (GEF) 3 | -193994 (-) | SPATA12|spermatogenesis associated 12 | 64598 (+) |
| chr10:100537960..100538129 | 5.026 | HPS1|Hermansky-Pudlak syndrome 1 | -331333 (-) | HPSE2|heparanase 2 | 457550 (-) |
| chr12:38248572..38248646 | 5.021 | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | -4073136 (+) | ALG10B|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast) | 462016 (+) |
| chr12:38029012..38029141 | 5.014 | ALG10|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog (S. pombe) | -3853603 (+) | ALG10B|asparagine-linked glycosylation 10, alpha-1,2-glucosyltransferase homolog B (yeast) | 681549 (+) |
| chr7:74379932..74380093 | 5.012 | GATSL1|GATS protein-like 1 GATSL2|GATS protein-like 2 | -932 (+) | WBSCR16|Williams-Beuren syndrome chromosome region 16 | 109590 (-) |
| chr14:22926110..22926252 | 5.009 | OR4E2|olfactory receptor, family 4, subfamily E, member 2 | -792859 (+) | 63168 (+) | |
| chrX:61881425..61881500 | 4.989 | ZXDA|zinc finger, X-linked, duplicated A | -3944503 (-) | SPIN4|spindlin family, member 4 | 689737 (-) |
| chr21:10819542..10819655 | 4.984 | None (None) | TPTE|transmembrane phosphatase with tensin homology | 171300 (-) | |
| chr8:101844715..101844847 | 4.978 | PABPC1|poly(A) binding protein, cytoplasmic 1 | -109815 (-) | YWHAZ|tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 91743 (-) |
| chr9:72653141..72653320 | 4.963 | C9orf135|chromosome 9 open reading frame 135 | -217494 (+) | MAMDC2|MAM domain containing 2 | 5277 (+) |
| chr16:35281331..35281415 | 4.950 | LOC653550|TP53-target gene 3 protein-like TP53TG3B|TP53 target 3B TP53TG3|TP53 target 3 | -2019240 (+) | SHCBP1|SHC SH2-domain binding protein 1 | 11373899 (-) |
| chr10:29798240..29798347 | 4.941 | SVIL|supervillin | -43742 (-) | 125568 (-) | |
| chr18:39971816..39971884 | 4.941 | PIK3C3|phosphoinositide-3-kinase, class 3 | -436617 (+) | RIT2|Ras-like without CAAX 2 | 723719 (-) |
| chr16:33391806..33391926 | 4.937 | LOC653550|TP53-target gene 3 protein-like TP53TG3B|TP53 target 3B TP53TG3|TP53 target 3 | -129733 (+) | SHCBP1|SHC SH2-domain binding protein 1 | 13263406 (-) |
| chr4:106629840..106629956 | 4.916 | INTS12|integrator complex subunit 12 | -57 (-) | GSTCD|glutathione S-transferase, C-terminal domain containing | 75 (+) |
| chr4:144299743..144299876 | 4.916 | GAB1|GRB2-associated binding protein 1 | -41794 (+) | SMARCA5|SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 5 | 134919 (+) |
| chr3:90473696..90473799 | 4.906 | EPHA3|EPH receptor A3 | -1316897 (+) | PROS1|protein S (alpha) | 3218929 (-) |
| chr7:30334573..30334686 | 4.905 | ZNRF2|zinc and ring finger 2 | -9973 (+) | NOD1|nucleotide-binding oligomerization domain containing 1 | 183692 (-) |
| chrX:113445887..113445953 | 4.899 | AMOT|angiomotin | -1361858 (-) | HTR2C|5-hydroxytryptamine (serotonin) receptor 2C | 372652 (+) |
| chr19:27789636..27789737 | 4.871 | ZNF254|zinc finger protein 254 | -3519717 (+) | 494702 (+) | |
| chrUn_gl000216:75741..75921 | 4.864 | None | None (None) | None | None (None) |
| chr19:27897234..27897334 | 4.863 | ZNF254|zinc finger protein 254 | -3627315 (+) | 387104 (+) | |
| chr3:7145435..7145500 | 4.856 | GRM7|glutamate receptor, metabotropic 7 | -242569 (+) | LOC100288428|uncharacterized LOC100288428 | 1397863 (-) |
| chr9:136429178..136429322 | 4.855 | ADAMTSL2|ADAMTS-like 2 | -29325 (+) | FAM163B|family with sequence similarity 163, member B | 16105 (-) |
| chr21:10736138..10736247 | 4.850 | None (None) | TPTE|transmembrane phosphatase with tensin homology | 254706 (-) |